
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,283,228 – 17,283,318 |
| Length | 90 |
| Max. P | 0.996914 |

| Location | 17,283,228 – 17,283,318 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.88 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
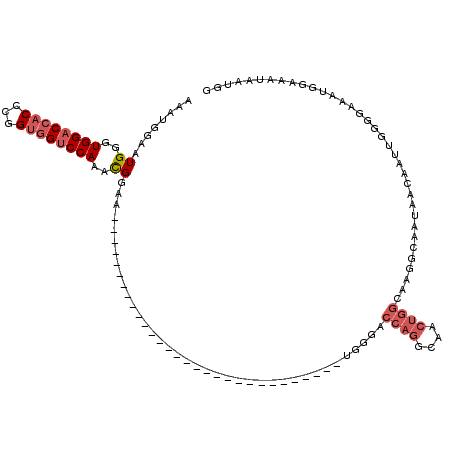
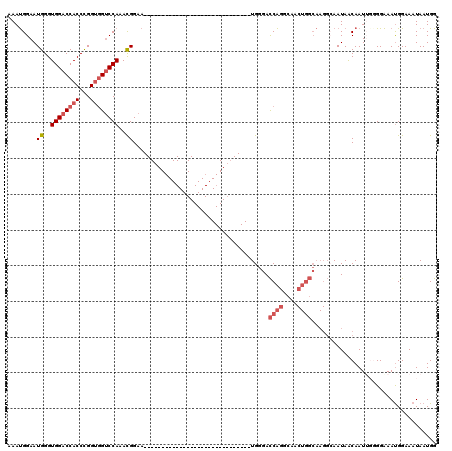
>2L_DroMel_CAF1 17283228 90 + 22407834 AAAUGGAAUGGGUGGACCACCUGGUGGUCCAAACGGAA------------------------------UGGGACCAGGCAACUGGCAAGGCAAUAACAAUUGGGGAAAUGGAAAUAAUGG ........((..((((((((...))))))))..))...------------------------------...(.((((....)))))....((((....)))).................. ( -23.60) >DroSec_CAF1 122330 87 + 1 AAAUGGAAUGGGUGGACCACCCGGUGGUCCAAACGGAA------------------------------UGGGACCAGGCAACUGGCAAGGCAAUAGCAAU---GGAAAUGGAAAUAAUGG ........((..((((((((...))))))))..))...------------------------------.....((((....))))((..((....))..)---)................ ( -24.60) >DroSim_CAF1 126137 87 + 1 AAAUGGAAUGGGUGGACCACCCGGUGGUCCAAACGGAA------------------------------UGGGACCAGGCAACUGGCAAGGCAAUAACAAU---GGAAACGGAAAUAAUGG ........((..((((((((...))))))))..))...------------------------------...(.((((....))))).............(---(....)).......... ( -24.90) >DroEre_CAF1 121502 120 + 1 AAAUGGAAUGGGUGGACCACCCGGCAGGCCAAAUGGAAUGGGUGGACCACCUGGUGGUCCAAACGGAAUGGGACCAGGCAACUGGCAAGGAAAUAACAAUUGGGGAAAUGGAAAUAAUGG ...((((.(((((((.(((((((.....((....))..))))))).)))))))....))))..........(.((((....))))).................................. ( -38.80) >DroYak_CAF1 123752 78 + 1 UAACGGAAUGGGUGGACCACCUGGUGGUCCAAACGGAA------------------------------UGGGAUCGGGC------------AAUAACAAUUGGGGGAAUGGAAAUAAUGG ...((...((..((((((((...))))))))..))...------------------------------))........(------------(((....)))).................. ( -16.90) >consensus AAAUGGAAUGGGUGGACCACCCGGUGGUCCAAACGGAA______________________________UGGGACCAGGCAACUGGCAAGGCAAUAACAAUUGGGGAAAUGGAAAUAAUGG ........((..((((((((...))))))))..))......................................((((....))))................................... (-15.64 = -16.88 + 1.24)

| Location | 17,283,228 – 17,283,318 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -12.58 |
| Energy contribution | -13.46 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
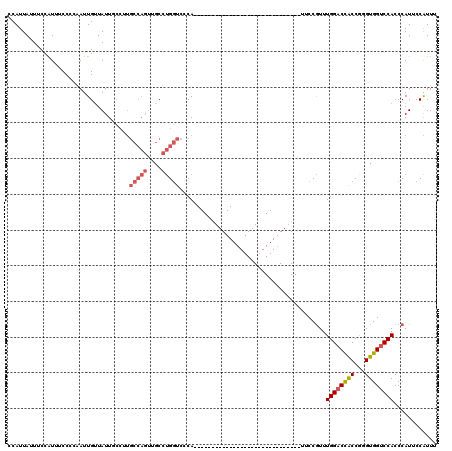
>2L_DroMel_CAF1 17283228 90 - 22407834 CCAUUAUUUCCAUUUCCCCAAUUGUUAUUGCCUUGCCAGUUGCCUGGUCCCA------------------------------UUCCGUUUGGACCACCAGGUGGUCCACCCAUUCCAUUU ..................................(((((....)))))....------------------------------....(..((((((((...))))))))..)......... ( -18.50) >DroSec_CAF1 122330 87 - 1 CCAUUAUUUCCAUUUCC---AUUGCUAUUGCCUUGCCAGUUGCCUGGUCCCA------------------------------UUCCGUUUGGACCACCGGGUGGUCCACCCAUUCCAUUU .................---...((....))...(((((....)))))....------------------------------....(..((((((((...))))))))..)......... ( -19.10) >DroSim_CAF1 126137 87 - 1 CCAUUAUUUCCGUUUCC---AUUGUUAUUGCCUUGCCAGUUGCCUGGUCCCA------------------------------UUCCGUUUGGACCACCGGGUGGUCCACCCAUUCCAUUU .................---..............(((((....)))))....------------------------------....(..((((((((...))))))))..)......... ( -18.50) >DroEre_CAF1 121502 120 - 1 CCAUUAUUUCCAUUUCCCCAAUUGUUAUUUCCUUGCCAGUUGCCUGGUCCCAUUCCGUUUGGACCACCAGGUGGUCCACCCAUUCCAUUUGGCCUGCCGGGUGGUCCACCCAUUCCAUUU ..................((((((.((......)).))))))..((((((.(......).))))))...(((((.((((((...((....))......)))))).))))).......... ( -33.80) >DroYak_CAF1 123752 78 - 1 CCAUUAUUUCCAUUCCCCCAAUUGUUAUU------------GCCCGAUCCCA------------------------------UUCCGUUUGGACCACCAGGUGGUCCACCCAUUCCGUUA .............................------------...........------------------------------....(..((((((((...))))))))..)......... ( -13.30) >consensus CCAUUAUUUCCAUUUCCCCAAUUGUUAUUGCCUUGCCAGUUGCCUGGUCCCA______________________________UUCCGUUUGGACCACCGGGUGGUCCACCCAUUCCAUUU ..................................(((((....))))).........................................((((((((...))))))))............ (-12.58 = -13.46 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:55 2006