
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,264,698 – 17,264,803 |
| Length | 105 |
| Max. P | 0.989488 |

| Location | 17,264,698 – 17,264,803 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
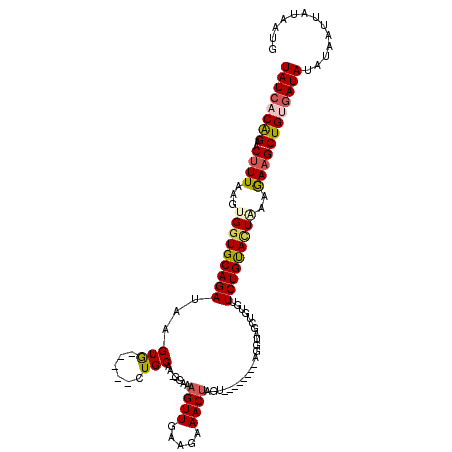
>2L_DroMel_CAF1 17264698 105 + 22407834 UAUCACAGAACUUUGAGUGUUGCAGAUAAGUA-----CUGCAA-AAAAAGUUGAAGAAACUAGU--------CGGUAGCUGUGUUCUGCACUGAAAAAGCUGAGAUAUAUAAUUAU-AUG ((((.(((..(((..(.(.((((((.......-----))))))-....).)..))).......(--------((((.((........))))))).....))).)))).........-... ( -23.30) >DroSec_CAF1 107337 108 + 1 UAUCACAGAACUUUAAGUGGUGCAGAUAAGUG-----CUGCGAUGGAAAGUUGAAGCAACUAGU-------UCGGUAGCUGUGUUCUGCACUAAGGAAGCUGUAAUAUAUAAUUACAAUG (((.((((..((((...(((((((((.....(-----((((..((((.(((((...)))))..)-------)))))))).....)))))))))..)))))))).)))............. ( -31.20) >DroSim_CAF1 112413 103 + 1 UAUCACAGAACUUUAAGUGGUGCAGAUAAGUG-----CUGCAA-GGAAAGUUGAAGCAACUAGU-------GAGGUAGCUGUGUUCUGUACUAAAGAAGCUGUGAUAUAUAAUU----UU ((((((((..((((...(((((((((.....(-----((((..-....(((((...)))))...-------...))))).....)))))))))..)))))))))))).......----.. ( -34.20) >DroEre_CAF1 106668 111 + 1 UAUCACGGAACUUUAAGUGGUGCAGAUAAGUG-----CUGCAA-GUAGAGUUGAAGAAACUAUAGCUGCUAUAGGUACCUGUGUUCUGUAUUCAAGAAGCUGUGAUACAUUA---AACUG (((((((((((((((.(..(..(......)..-----)..)..-.)))))))...(((..(((((..((.((((....)))))).))))))))......)))))))).....---..... ( -29.10) >DroYak_CAF1 109471 108 + 1 UAUCACAGAACUUUAAGUGGUGCAGAUAAGUACCAUACUGCAA-GUAAUGUUGAAGAAACU-----------AGGUACCUGUGUUCUGUAUUCAAGAUGCUGAGAUACAUAAUAAUAUUG (((((((((((.....(((((((......)))))))...(((.-(((..(((.....))).-----------...))).))))))))))..(((......)))))))............. ( -24.70) >consensus UAUCACAGAACUUUAAGUGGUGCAGAUAAGUG_____CUGCAA_GGAAAGUUGAAGAAACUAGU________AGGUAGCUGUGUUCUGUACUAAAGAAGCUGUGAUAUAUAAUUAUAAUG ((((((((..((((...(((((((((...(((......))).......((((.....)))).......................)))))))))..))))))))))))............. (-17.68 = -18.24 + 0.56)

| Location | 17,264,698 – 17,264,803 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
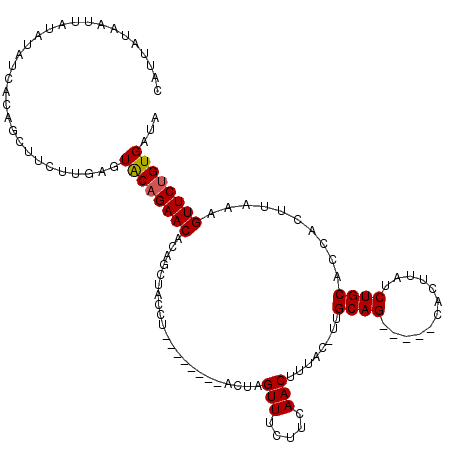
>2L_DroMel_CAF1 17264698 105 - 22407834 CAU-AUAAUUAUAUAUCUCAGCUUUUUCAGUGCAGAACACAGCUACCG--------ACUAGUUUCUUCAACUUUUU-UUGCAG-----UACUUAUCUGCAACACUCAAAGUUCUGUGAUA ...-.........((((.(((..((((.((((..(((...(((((...--------..)))))..)))........-((((((-----.......)))))))))).))))..))).)))) ( -20.70) >DroSec_CAF1 107337 108 - 1 CAUUGUAAUUAUAUAUUACAGCUUCCUUAGUGCAGAACACAGCUACCGA-------ACUAGUUGCUUCAACUUUCCAUCGCAG-----CACUUAUCUGCACCACUUAAAGUUCUGUGAUA .............(((((((((((.....(((((((.....(((..(((-------...(((((...))))).....))).))-----).....)))))))......)))..)))))))) ( -23.30) >DroSim_CAF1 112413 103 - 1 AA----AAUUAUAUAUCACAGCUUCUUUAGUACAGAACACAGCUACCUC-------ACUAGUUGCUUCAACUUUCC-UUGCAG-----CACUUAUCUGCACCACUUAAAGUUCUGUGAUA ..----.......((((((((...(((((((...(((..((((((....-------..)))))).)))........-.(((((-----.......)))))..)).)))))..)))))))) ( -23.20) >DroEre_CAF1 106668 111 - 1 CAGUU---UAAUGUAUCACAGCUUCUUGAAUACAGAACACAGGUACCUAUAGCAGCUAUAGUUUCUUCAACUCUAC-UUGCAG-----CACUUAUCUGCACCACUUAAAGUUCCGUGAUA .....---.....((((((.(.((((.......)))))...(((.......((((.((.((((.....)))).)).-)))).(-----((......))))))............)))))) ( -17.20) >DroYak_CAF1 109471 108 - 1 CAAUAUUAUUAUGUAUCUCAGCAUCUUGAAUACAGAACACAGGUACCU-----------AGUUUCUUCAACAUUAC-UUGCAGUAUGGUACUUAUCUGCACCACUUAAAGUUCUGUGAUA .....(((..((((......))))..))).((((((((.((((((...-----------.(((.....)))..)))-))).....((((.(......).))))......))))))))... ( -19.60) >consensus CAUUAUAAUUAUAUAUCACAGCUUCUUGAGUACAGAACACAGCUACCU________ACUAGUUUCUUCAACUUUAC_UUGCAG_____CACUUAUCUGCACCACUUAAAGUUCUGUGAUA ..............................((((((((......................(((.....)))........((((............))))..........))))))))... (-12.04 = -12.00 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:48 2006