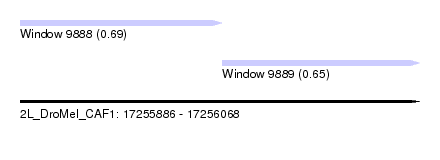
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,255,886 – 17,256,068 |
| Length | 182 |
| Max. P | 0.694754 |

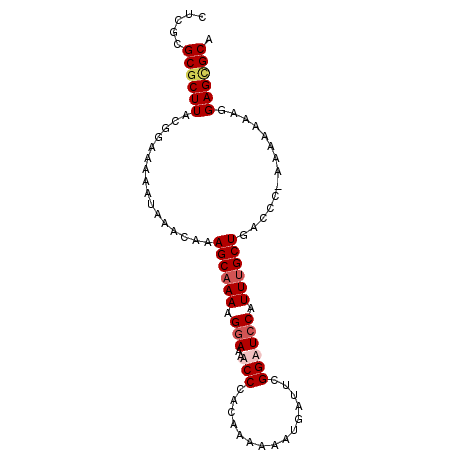
| Location | 17,255,886 – 17,255,978 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -11.69 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17255886 92 + 22407834 CCAUUUUCCGCCGCACCCA----------AAAGGC-AAAGCAAAACUGGAUGAACGCAUAAACAAAUAUGCCAAAAACAUAGUA-AAAAAAUUGUGUGCAAAAU .........(((.......----------...)))-...((..............(((((......))))).....(((((((.-.....)))))))))..... ( -15.00) >DroSec_CAF1 98632 92 + 1 CCAUUUUGCAACGCACCCA----------A-AGGC-AAAGCCAAACUGGAUGAACGCAUAAACAAAUAUGCCAAAAACAUAGUAGAAAAAACUGUGUGCAAAAU ..(((((((....((.(((----------.-.(((-...)))....))).))...(((((......))))).....(((((((.......)))))))))))))) ( -20.80) >DroSim_CAF1 103687 92 + 1 CCAUUUUGCAACGCACCCA----------A-AGGC-AAAGCAAAACUGGAUGAACGCAUAAACAAAUAUGCCAAAAACAUAGUAGAAAAAACUGUGUGCAAAAU (((((((((...((.....----------.-..))-...)))))).)))......(((((......))))).....(((((((.......)))))))....... ( -19.20) >DroEre_CAF1 97707 92 + 1 CCAUUUUCCAGCGCGGCGA----------C-AGGC-AAAGCAAAACUGGCUGAACGCAUAAACAAAUAUGCCAAAAACAUAGUAGAAAAAACUGUGUGGAAAAU ..(((((((.(..((((..----------(-((..-.........)))))))..)(((((......))))).....(((((((.......)))))))))))))) ( -22.10) >DroYak_CAF1 99490 92 + 1 CCAUUUUCCACUGCGCCCA----------C-AGGC-GAAGCAAAACUGGCUGAACGCAUAAACAAAUAUGCCAAAAACAUAGUAGAAAAAACUGUGUGGAAAAU ..(((((((....((((..----------.-.)))-).(((.......)))....(((((......))))).....(((((((.......)))))))))))))) ( -22.90) >DroAna_CAF1 168746 100 + 1 CCAUUUUCGCCCCAGCCCAGUCGGAUUUAA-AUCCGAGACCAAAACUUGC-CAACGCAUAAACAAAUAUGCCAAAAAA--AAUACAAAAAAAUGUGUGGAAAAU ...(((((((..........((((((....-)))))).............-....(((((......))))).......--...............))))))).. ( -16.00) >consensus CCAUUUUCCACCGCACCCA__________A_AGGC_AAAGCAAAACUGGAUGAACGCAUAAACAAAUAUGCCAAAAACAUAGUAGAAAAAACUGUGUGCAAAAU ..(((((((..............................................(((((......))))).....(((((((.......)))))))))))))) (-11.69 = -12.25 + 0.56)

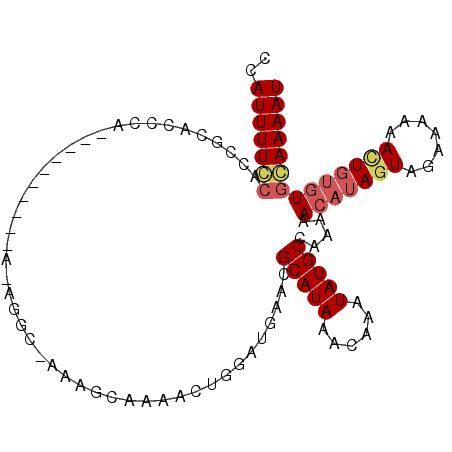
| Location | 17,255,978 – 17,256,068 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -13.85 |
| Energy contribution | -14.97 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17255978 90 + 22407834 CUCGCGCGCUUUCGGAAAAAUAAACAAAGCUAAAGGAAAACCCACAAAAAAUGAUUUGGAUCCAUUUGCUGACCC-AAAAAAAGGAGCGCA ...(((((((((.(..........))))))....((.....))...........(((((.((........)).))-))).......)))). ( -16.50) >DroSec_CAF1 98724 91 + 1 CUCGCGCUCUUACGGAAAAAUAAACAAAGCAAAAGGAAAUCCCACAAGAAAUGAUUCGGAUCCAUUUGCUGACCCCAAAAAAAGGAGCGCA ...((((((((..(..........)..((((((.(((..(((....(....).....)))))).))))))............)))))))). ( -23.60) >DroSim_CAF1 103779 89 + 1 CUCGCGCGCUUACGGAAAAAUAAACAAAGCAAAAGGAAAUCCCACAAAAAAUGAUUCGGAUCCAUUUGCUGACCC--AAAAAAGGAGCGCA .....(((((.................((((((.(((..(((...............)))))).))))))...((--......))))))). ( -19.86) >DroYak_CAF1 99582 90 + 1 CUCGCGCGCUUACGGCCAAACAAACAAAGCAAAGGCAAAACCCACAAAAAAUGAUUCGGAUCCAUUUGCUGAUCC-AAAAAAAGGAGUGCA .....((((((...(((................))).....................(((((........)))))-........)))))). ( -17.99) >consensus CUCGCGCGCUUACGGAAAAAUAAACAAAGCAAAAGGAAAACCCACAAAAAAUGAUUCGGAUCCAUUUGCUGACCC_AAAAAAAGGAGCGCA .....((((((................((((((.(((..(((...............)))))).))))))..............)))))). (-13.85 = -14.97 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:44 2006