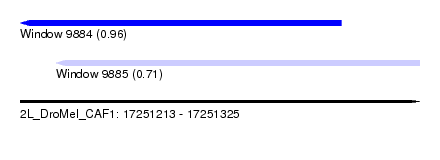
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,251,213 – 17,251,325 |
| Length | 112 |
| Max. P | 0.964575 |

| Location | 17,251,213 – 17,251,303 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -17.35 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
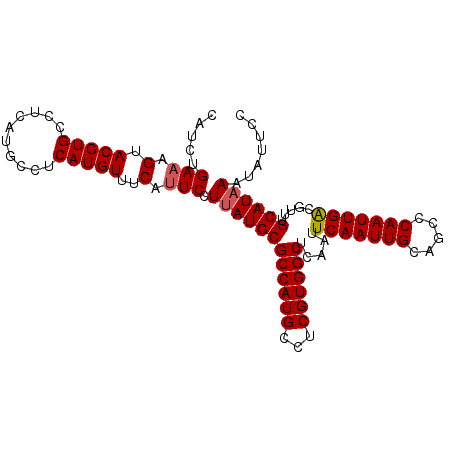
>2L_DroMel_CAF1 17251213 90 - 22407834 GCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCCCUUUCGACUC .................((((((((((((...)))))).....(((((((.....))))))).....))))))................. ( -18.50) >DroPse_CAF1 161260 79 - 1 -----------CGUGCCUCAUGCCCCAUGCCUCAUGGCUCAAUUCAAUUGCAGGCCAAUUGACGUUUGCAUAAAUAUUGUAUUCCCCCUC -----------.................(((....))).((((.....(((((((........))))))).....))))........... ( -13.10) >DroSim_CAF1 98921 90 - 1 GCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCCCUUUCGACUC .................((((((((((((...)))))).....(((((((.....))))))).....))))))................. ( -18.50) >DroEre_CAF1 93208 90 - 1 GCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCUCAAUUGGCGUUUGCAUAAAUAUUCCCUUUCGACUC .................(((((((..(((((...(((((((((...)))).))).))...))))).)))))))................. ( -19.10) >DroYak_CAF1 94889 90 - 1 GCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCCCUUUCGACUC .................((((((((((((...)))))).....(((((((.....))))))).....))))))................. ( -18.50) >DroAna_CAF1 164395 85 - 1 GCCUCAUGUUUCAUUCCUCAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAU-----UUCGACUC ...................((((((((((...)))))).....(((((((.....))))))).....))))......-----........ ( -16.40) >consensus GCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCCCUUUCGACUC .................((((((((((((...)))))).....(((((((.....))))))).....))))))................. (-16.38 = -16.60 + 0.22)



| Location | 17,251,223 – 17,251,325 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -19.75 |
| Energy contribution | -19.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17251223 102 - 22407834 CAUCUGAAAGUACGUGCCUCGUGCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCC ....(((..(((((.....))))).)))...........((((((((((((...)))))).....(((((((.....))))))).....))))))....... ( -24.10) >DroSec_CAF1 94092 102 - 1 CGUCUGAAAGUACGUGCCUCAUGCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCC .(..((((...(((((..........)))))))))..).((((((((((((...)))))).....(((((((.....))))))).....))))))....... ( -21.10) >DroSim_CAF1 98931 102 - 1 CAUCUGAAAGUACGUGCCUCGUGCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCC ....(((..(((((.....))))).)))...........((((((((((((...)))))).....(((((((.....))))))).....))))))....... ( -24.10) >DroEre_CAF1 93218 102 - 1 CAUCUGAAAGUACGUGCCUCAUGCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCUCAAUUGGCGUUUGCAUAAAUAUUCC ....((((...(((((..........)))))))))....(((((((..(((((...(((((((((...)))).))).))...))))).)))))))....... ( -21.60) >DroYak_CAF1 94899 102 - 1 CAUCUGAAAGUACGUGCCGCAUGCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCC ....((((...(((((..(....)..)))))))))....((((((((((((...)))))).....(((((((.....))))))).....))))))....... ( -22.40) >DroAna_CAF1 164403 98 - 1 CAUCCGA-AGUACGUGCCUCUUGCCUCAUGUUUCAUUCCUCAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAU--- .....((-(..(((((..........)))))....)))...((((((((((...)))))).....(((((((.....))))))).....))))......--- ( -19.30) >consensus CAUCUGAAAGUACGUGCCUCAUGCCUCAUGUUUCAUUCCUUAUGCGCCAUGCCUCGUGGCUCAAUUCAAUUGCAGCCCAAUUGACGUUUGCAUAAAUAUUCC .....(((.(.(((((..........)))))..).))).((((((((((((...)))))).....(((((((.....))))))).....))))))....... (-19.75 = -19.95 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:40 2006