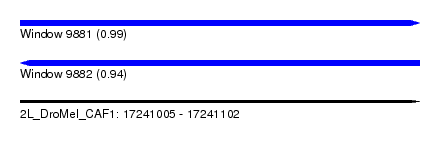
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,241,005 – 17,241,102 |
| Length | 97 |
| Max. P | 0.992430 |

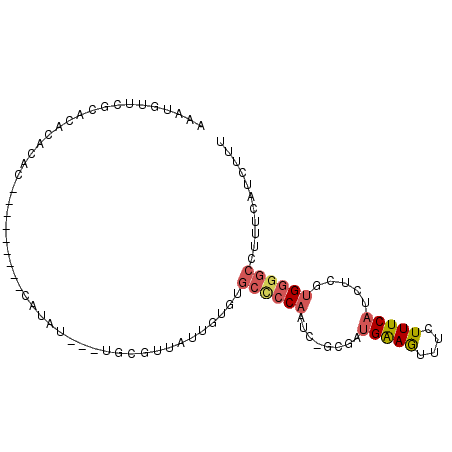
| Location | 17,241,005 – 17,241,102 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -7.71 |
| Energy contribution | -8.24 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17241005 97 + 22407834 AAAGAUGAAAGGCCCCAAGAGAUGAAAGAAACUUCAUCGC-GAUUGGGGCACACAAUAACGCAGCGACAUGUGUGUGUGUGAGUGUGUGCGAACAUUU ...........(((((((..((((((......))))))..-..)))))))(((((...(((((......))))).)))))((((((......)))))) ( -34.90) >DroPse_CAF1 148380 90 + 1 AAAGAUGAAACGUGGCA-AAGUUGAAAGAAAUUCCACGAAAAAAUGGAGGGGAAAAAAAGCCUCGAAUAAAA------A-AAAGAUGUGUGAAUAUUU ..........(((((.(-(..((....))..)))))))..........((((........))))........------.-..((((((....)))))) ( -13.60) >DroSec_CAF1 84107 79 + 1 AAAGAUGAAAGGCCCCACGAGAUGAAAGAAACUUCAUCGC-GAUUGGGGCACACAAUAACGC------------------GUGUGUGUGCGAACAUUU ..(((((....((((((((.((((((......)))))).)-)..))))))(((((.......------------------.))))).......))))) ( -26.00) >DroSim_CAF1 86971 79 + 1 AAAGAUGAAAGGCCCCACGAGAUGAAAGAAACUUCAUCGC-GAUUGGGGCACACAAUAACGC------------------GUGUGUGUGCGAACAUUU ..(((((....((((((((.((((((......)))))).)-)..))))))(((((.......------------------.))))).......))))) ( -26.00) >DroEre_CAF1 83646 89 + 1 AAAGAUGAAAGGCCCCAGGAGAUGAAAGAAACUUCUGCAC-AAU-GGGGCACACAAUAAUACACGUAUGUGU------G-GUGUGUGUGCGAGCAUUU ..(((((.....(((((((((.(......).))))))...-...-)))(((((((....(((((....))))------)-...)))))))...))))) ( -26.20) >DroYak_CAF1 84792 89 + 1 AAAGAUGAAAGGCCCCAAGAGAUGAAAGAAACUUCAGCAC-GAUUGGAGCGAACAAUAACGCAGAGAUAUGC--------GUGUGUGUGCGAGCAUUU ..(((((...(((.((((..(.((((......))))...)-..)))).))..(((...(((((......)))--------))...))).)...))))) ( -20.90) >consensus AAAGAUGAAAGGCCCCAAGAGAUGAAAGAAACUUCAUCAC_GAUUGGGGCACACAAUAACGCA___AUAUG_________GUGUGUGUGCGAACAUUU ...........((((((.....((((......))))........))))))..................................((((....)))).. ( -7.71 = -8.24 + 0.53)

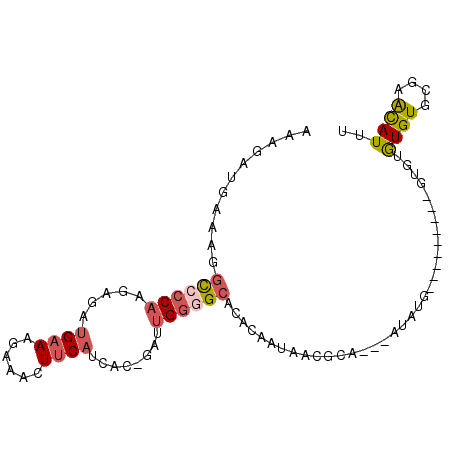
| Location | 17,241,005 – 17,241,102 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -7.59 |
| Energy contribution | -8.20 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17241005 97 - 22407834 AAAUGUUCGCACACACUCACACACACACAUGUCGCUGCGUUAUUGUGUGCCCCAAUC-GCGAUGAAGUUUCUUUCAUCUCUUGGGGCCUUUCAUCUUU ........(((((((.(.((.(((((....)).).)).)).).)))))))(((((..-(.(((((((....))))))).))))))............. ( -23.70) >DroPse_CAF1 148380 90 - 1 AAAUAUUCACACAUCUUU-U------UUUUAUUCGAGGCUUUUUUUCCCCUCCAUUUUUUCGUGGAAUUUCUUUCAACUU-UGCCACGUUUCAUCUUU ..................-.------........((((..........))))........(((((...............-..))))).......... ( -8.03) >DroSec_CAF1 84107 79 - 1 AAAUGUUCGCACACACAC------------------GCGUUAUUGUGUGCCCCAAUC-GCGAUGAAGUUUCUUUCAUCUCGUGGGGCCUUUCAUCUUU ............(((((.------------------.......)))))((((((..(-(.(((((((....))))))).))))))))........... ( -23.20) >DroSim_CAF1 86971 79 - 1 AAAUGUUCGCACACACAC------------------GCGUUAUUGUGUGCCCCAAUC-GCGAUGAAGUUUCUUUCAUCUCGUGGGGCCUUUCAUCUUU ............(((((.------------------.......)))))((((((..(-(.(((((((....))))))).))))))))........... ( -23.20) >DroEre_CAF1 83646 89 - 1 AAAUGCUCGCACACACAC-C------ACACAUACGUGUAUUAUUGUGUGCCCC-AUU-GUGCAGAAGUUUCUUUCAUCUCCUGGGGCCUUUCAUCUUU ....(((((((((((...-.------((((....)))).....)))))))...-...-...(((.((..........)).)))))))........... ( -19.90) >DroYak_CAF1 84792 89 - 1 AAAUGCUCGCACACACAC--------GCAUAUCUCUGCGUUAUUGUUCGCUCCAAUC-GUGCUGAAGUUUCUUUCAUCUCUUGGGGCCUUUCAUCUUU ........(.(((...((--------(((......)))))...))).)(((((((..-(.(.(((((....))))).).))))))))........... ( -17.80) >consensus AAAUGUUCGCACACACAC_________CAUAU___UGCGUUAUUGUGUGCCCCAAUC_GCGAUGAAGUUUCUUUCAUCUCGUGGGGCCUUUCAUCUUU ................................................((((((........(((((....))))).....))))))........... ( -7.59 = -8.20 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:36 2006