| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,829,996 – 1,830,093 |
| Length | 97 |
| Max. P | 0.540377 |

| Location | 1,829,996 – 1,830,093 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -17.33 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
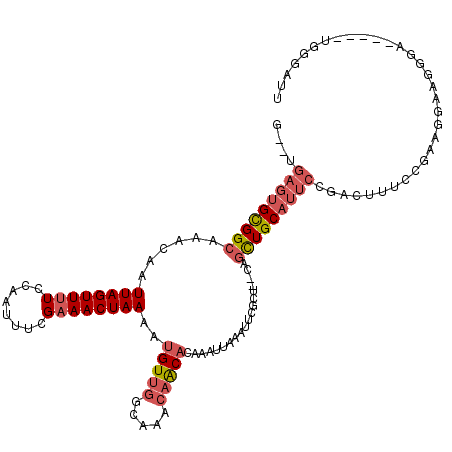
>2L_DroMel_CAF1 1829996 97 - 22407834 G--UGAGUGCGGCAAACAAUUAGUUUUCCAAUUUCGAAACUAAAAUGUUGGCAAACAACACAAAUUAAAUUCACU--CAGUUGCAUUCCGACUUUUCGAAG------------------- .--.(((((((((......((((((((........))))))))..(((((.....)))))...............--..)))))))))(((....)))...------------------- ( -22.30) >DroSec_CAF1 50036 111 - 1 G--UGAGUGCGGCAAACAAUUAGUUUUCCAAUUUCGAAACUAAAAUGUUGGCAAACAACACAAAUUAAAUUCGCU--CAGCUGCAUUCCGACUUUCCGAAGGAAGGAA-----UGGGAUU (--.(((((((((......((((((((........))))))))..(((((.....)))))...............--..))))))))))..((((((...))))))..-----....... ( -29.60) >DroSim_CAF1 30592 111 - 1 G--UGAGUGCGGCAAACAAUUAGUUUUCCAAUUUCGAAACUAAAAUGUCGGCAAACAACACAAAUUAAAUUCGCU--CAGCUGCAUUCCGAUUUUCCGAAGGAAGGGA-----UGGGAUU (--.(((((((((......((((((((........))))))))..(((.(.....).)))...............--..))))))))))....((((.......))))-----....... ( -23.00) >DroPer_CAF1 47272 115 - 1 GGAUGUGUGUGGCAAACAAUUAGUUUUCCAAUUU-GAAACUAAAAUGUUUGCAAACAGCUCAAAUAAAAUUCCUUCCCAUCUGC----CGUCUCUGCCAAAUAGUGGAUUCAGUGGAAUG .((..(((...(((((((.((((((((.......-))))))))..)))))))..)))..))..................((..(----.(..((..(......)..))..).)..))... ( -25.20) >DroEre_CAF1 38512 112 - 1 G--UGAGUGUGGCAAACAAUUAGUUUUCCAAUUUCGAAACUAAAAUGUUGGCAAACAACACAAAUUAAAUUCGCC-CGAGCUGCAUUCUGAUUUUCCGAAGGAGGGGA-----AAGGAUU .--.(((((..((......((((((((........))))))))..(((((.....)))))...............-...))..)))))...((..((......))..)-----)...... ( -27.00) >DroYak_CAF1 15024 112 - 1 G--UGAGUGCGGCAAACAAUUAGUUUUCCAAUUUGGCAACUAAAAUGUUGGCAAACAACACAAAUUAAAUUCGCC-UGAGUUGCAUUCCGACUUUGCGAUGGAAGGGA-----AAGGAUU (--((.....((.((((.....)))).))..((((.((((......)))).))))...)))........(((.((-(..((((((.........))))))...)))))-----)...... ( -26.80) >consensus G__UGAGUGCGGCAAACAAUUAGUUUUCCAAUUUCGAAACUAAAAUGUUGGCAAACAACACAAAUUAAAUUCGCU__CAGCUGCAUUCCGACUUUCCGAAGGAAGGGA_____UGGGAUU ....(((((((((......((((((((........))))))))..(((((.....)))))...................)))))))))................................ (-17.33 = -18.25 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:07 2006