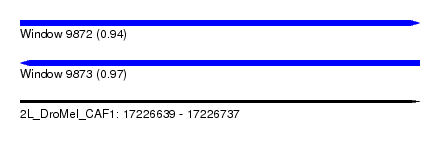
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,226,639 – 17,226,737 |
| Length | 98 |
| Max. P | 0.971211 |

| Location | 17,226,639 – 17,226,737 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -14.33 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
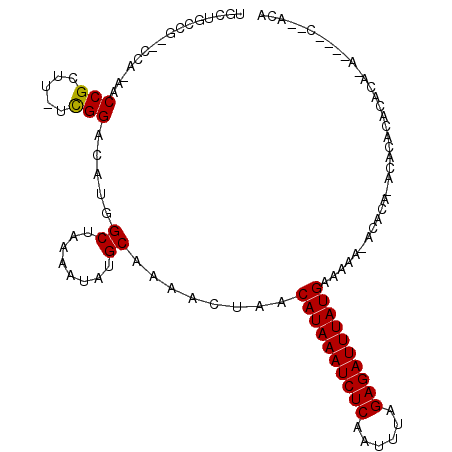
>2L_DroMel_CAF1 17226639 98 + 22407834 CG------------AGUGUGUGUGUGUGU-UUUUUCAUAAAUCUCCAAAUUGAGAUUUAUGUUAGUUUUGCAUAUUUUAGCCAUGUCCGU-GAGCGGUU-AGCAGCGCCCGCA ..------------.(((.((((((((((-.((..((((((((((......))))))))))..))....))))))....((.....(((.-...)))..-.)).)))).))). ( -27.50) >DroVir_CAF1 166582 100 + 1 UGU--G----UGUGUGUGUGUGCAU---U-UUUUUCAUAAAUCUCUAAAUUGAGAUUUAUGUUAGUUUUGCAUAUUUUAGCCAUGUCCGAAAAGCGGUU-UGG--CGGCAGCA ...--.----(((.((((((((((.---.-.((..((((((((((......))))))))))..))...)))))))....((((...(((.....)))..-)))--).)))))) ( -32.60) >DroGri_CAF1 196983 107 + 1 UUU--GCUGUGCUGUGUGUGCGUGUGUGU-UUUUUCAUAAAUCUCUAAAUUGAGAUUUAUGAUAGUUUUGCAUAUUUUAGCCAUGUCCGAGAAGCGGUU-UGG--CGGCAACA .((--((((..(.....)..)(((((((.-....(((((((((((......)))))))))))......)))))))....((((...(((.....)))..-)))--)))))).. ( -35.90) >DroEre_CAF1 69520 103 + 1 CGUACG----UGUGUGUGUG----UGUGUGUUUUUCAUAAAUCUCCAAAUUGAGAUUUAUGUUAGUUUUGCAUAUUUUAGCCAUGUCCGA-GAGCGGUU-GGCAUCGCCGGCA ((.(((----((.((..(((----((((...((..((((((((((......))))))))))..))...)))))))....))))))).)).-.....(((-(((...)))))). ( -32.10) >DroWil_CAF1 84959 89 + 1 U--------------GUGUG----UGCGAGUUUUUCAUAAAUCUCUAAAUUGAGAUUUAUGUUAGUUUUGCAUAUUUUAGCCAUGUCCAA-AAGCGGUU-UGG--CGGCC--A .--------------..(((----((((((.((..((((((((((......))))))))))..)).)))))))))....((((...((..-....))..-)))--)....--. ( -30.40) >DroMoj_CAF1 145507 104 + 1 CGU--G----UAUGUGUGUGUGUAUGUAU-UUUUUCAUAAAUCUCUAAAUUGAGAUUUAUGUUAGUUUUGCAUAUUUUAGCCAUGUCCAAAAAGCGGUUUUGG--CGGCAUCA .(.--.----((((.((....(((((((.-.((..((((((((((......))))))))))..))...)))))))....))))))..).....((.((....)--).)).... ( -29.30) >consensus CGU__G____U_UGUGUGUGUGU_UGUGU_UUUUUCAUAAAUCUCUAAAUUGAGAUUUAUGUUAGUUUUGCAUAUUUUAGCCAUGUCCGA_AAGCGGUU_UGG__CGGCAGCA .......................(((((...((..((((((((((......))))))))))..))...))))).......(((...(((.....)))...))).......... (-14.33 = -15.83 + 1.50)


| Location | 17,226,639 – 17,226,737 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17226639 98 - 22407834 UGCGGGCGCUGCU-AACCGCUC-ACGGACAUGGCUAAAAUAUGCAAAACUAACAUAAAUCUCAAUUUGGAGAUUUAUGAAAAA-ACACACACACACACU------------CG ((((......(((-(.(((...-.)))...)))).......)))).......((((((((((......)))))))))).....-...............------------.. ( -21.22) >DroVir_CAF1 166582 100 - 1 UGCUGCCG--CCA-AACCGCUUUUCGGACAUGGCUAAAAUAUGCAAAACUAACAUAAAUCUCAAUUUAGAGAUUUAUGAAAAA-A---AUGCACACACACACA----C--ACA .......(--(((-..(((.....)))...)))).......((((.......((((((((((......)))))))))).....-.---.))))..........----.--... ( -22.64) >DroGri_CAF1 196983 107 - 1 UGUUGCCG--CCA-AACCGCUUCUCGGACAUGGCUAAAAUAUGCAAAACUAUCAUAAAUCUCAAUUUAGAGAUUUAUGAAAAA-ACACACACGCACACACAGCACAGC--AAA ((((...(--(((-..(((.....)))...))))...))))(((.......(((((((((((......)))))))))))....-........((.......))...))--).. ( -26.00) >DroEre_CAF1 69520 103 - 1 UGCCGGCGAUGCC-AACCGCUC-UCGGACAUGGCUAAAAUAUGCAAAACUAACAUAAAUCUCAAUUUGGAGAUUUAUGAAAAACACACA----CACACACACA----CGUACG .((((((...)))-..(((...-.)))....)))..................((((((((((......))))))))))...........----..........----...... ( -22.10) >DroWil_CAF1 84959 89 - 1 U--GGCCG--CCA-AACCGCUU-UUGGACAUGGCUAAAAUAUGCAAAACUAACAUAAAUCUCAAUUUAGAGAUUUAUGAAAAACUCGCA----CACAC--------------A (--((((.--(((-((.....)-))))....))))).....(((........((((((((((......))))))))))........)))----.....--------------. ( -22.89) >DroMoj_CAF1 145507 104 - 1 UGAUGCCG--CCAAAACCGCUUUUUGGACAUGGCUAAAAUAUGCAAAACUAACAUAAAUCUCAAUUUAGAGAUUUAUGAAAAA-AUACAUACACACACACAUA----C--ACG ....(((.--((((((.....))))))....)))..................((((((((((......)))))))))).....-...................----.--... ( -20.40) >consensus UGCUGCCG__CCA_AACCGCUU_UCGGACAUGGCUAAAAUAUGCAAAACUAACAUAAAUCUCAAUUUAGAGAUUUAUGAAAAA_ACACA_ACACACACACA_A____C__ACA ................(((.....))).....((........))........((((((((((......))))))))))................................... (-15.49 = -15.27 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:28 2006