| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,224,418 – 17,224,533 |
| Length | 115 |
| Max. P | 0.600528 |

| Location | 17,224,418 – 17,224,533 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -7.72 |
| Energy contribution | -10.32 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.30 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17224418 115 + 22407834 CUUUAGUGAAGG--GGCAAAUGAUGGGAUUUUCUUUUUGUGUGGUUAUCAAUAACAAAAUAAAUAAAUUCAAUUGGACUUAAAUGGUUGUUUUGUCACUUUUGCUAAUGGUGAGCUU ..((((..((((--((((((.....((((((...((((((.((.....))...)))))).....)))))).....((((.....))))..)))))).))))..)))).......... ( -23.50) >DroSec_CAF1 67630 113 + 1 CUUUAGUGAAGGGGGGCAAAUGAUGGGAUUUUCUUAUUGUGUGUUUAUCAAUAACAAAAU----AAAUUCAAUUGGACUUAAAUGGUUGUUUUGCCCCUUUUGCUAGAGGUGAGCUA (((((((.(((((((((((((((....(((((.((((((.........)))))).)))))----....)))....((((.....))))..)))))))))))))))))))........ ( -33.50) >DroSim_CAF1 69698 111 + 1 CUUUAGUGAAGG--GGCAAAUGAUGGGAUUUUCUUUUUGUGUGGUUAUCAAUAACAAAAU----AAAUUCAAUUGGACUUAAAUGGUUGUUUUGUCACUUUUGCUAGAGGUGAACUU ((((((..((((--((((((.....((((((...((((((.((.....))...)))))).----)))))).....((((.....))))..)))))).))))..))))))........ ( -28.20) >DroEre_CAF1 67299 95 + 1 CUUUAGUGGAGG--GGCAAAUGAUGGGAUUUUCUUUUUGUGUGGCUAC-AAUAGCGACAU----AAAUUCAAU-----------GGUGGCUUUUUCACCU----UUGAGGUGAACGA ((((((((((((--(((..((.((.(((((.......(((((.((((.-..)))).))))----)))))).))-----------.)).))))))))))..----.)))))....... ( -22.91) >DroYak_CAF1 68182 89 + 1 CUUUAGUGAAGG--GCAAAAUGAUGGGUUUAUCUUGUUGUGUGGUUACCAAUAACGAAAC----AAAUUCAAU-------------UGGUUUUGC---------UAAAGGUGAACAA ((((((..((.(--((...(..(..((.....))..)..)...)))((((((...(((..----...))).))-------------))))))..)---------)))))........ ( -19.40) >consensus CUUUAGUGAAGG__GGCAAAUGAUGGGAUUUUCUUUUUGUGUGGUUAUCAAUAACAAAAU____AAAUUCAAUUGGACUUAAAUGGUUGUUUUGUCACUUUUGCUAGAGGUGAACUA ((((((((((((...(((((..(..((.....))..)..)...((((....))))....................................))))..))))))))))))........ ( -7.72 = -10.32 + 2.60)

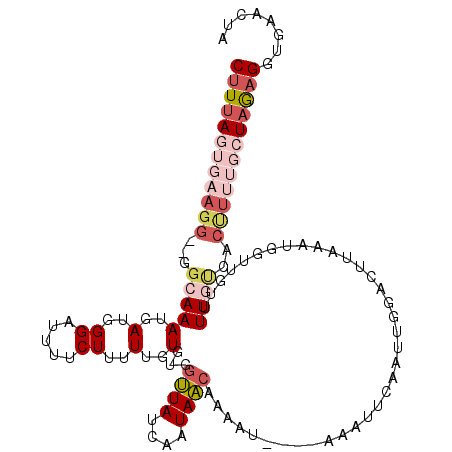
| Location | 17,224,418 – 17,224,533 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -13.70 |
| Consensus MFE | -0.04 |
| Energy contribution | 0.08 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.00 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17224418 115 - 22407834 AAGCUCACCAUUAGCAAAAGUGACAAAACAACCAUUUAAGUCCAAUUGAAUUUAUUUAUUUUGUUAUUGAUAACCACACAAAAAGAAAAUCCCAUCAUUUGCC--CCUUCACUAAAG .............(((((((((((((((.....((((((......)))))).......))))))))))................((........)).))))).--............ ( -10.90) >DroSec_CAF1 67630 113 - 1 UAGCUCACCUCUAGCAAAAGGGGCAAAACAACCAUUUAAGUCCAAUUGAAUUU----AUUUUGUUAUUGAUAAACACACAAUAAGAAAAUCCCAUCAUUUGCCCCCCUUCACUAAAG ..(((.......)))....((((((((......((((((......))))))..----(((((.((((((.........)))))).))))).......))))))))............ ( -21.50) >DroSim_CAF1 69698 111 - 1 AAGUUCACCUCUAGCAAAAGUGACAAAACAACCAUUUAAGUCCAAUUGAAUUU----AUUUUGUUAUUGAUAACCACACAAAAAGAAAAUCCCAUCAUUUGCC--CCUUCACUAAAG .............(((((((((((((((.((..((((((......))))))))----.))))))))))................((........)).))))).--............ ( -11.40) >DroEre_CAF1 67299 95 - 1 UCGUUCACCUCAA----AGGUGAAAAAGCCACC-----------AUUGAAUUU----AUGUCGCUAUU-GUAGCCACACAAAAAGAAAAUCCCAUCAUUUGCC--CCUCCACUAAAG ...(((...((((----.((((.......))))-----------.)))).(((----.(((.((((..-.)))).))).)))..)))................--............ ( -13.50) >DroYak_CAF1 68182 89 - 1 UUGUUCACCUUUA---------GCAAAACCA-------------AUUGAAUUU----GUUUCGUUAUUGGUAACCACACAACAAGAUAAACCCAUCAUUUUGC--CCUUCACUAAAG ........(((((---------(....((((-------------((((((...----..))))..))))))..........((((((.........)))))).--......)))))) ( -11.20) >consensus UAGUUCACCUCUAGCAAAAGUGACAAAACAACCAUUUAAGUCCAAUUGAAUUU____AUUUUGUUAUUGAUAACCACACAAAAAGAAAAUCCCAUCAUUUGCC__CCUUCACUAAAG ..............................................................((((....))))........................................... ( -0.04 = 0.08 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:26 2006