
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,210,295 – 17,210,420 |
| Length | 125 |
| Max. P | 0.997523 |

| Location | 17,210,295 – 17,210,390 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -20.45 |
| Energy contribution | -24.40 |
| Covariance contribution | 3.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17210295 95 + 22407834 AAUUUGGCCACGCCUAAUGACCAAAAAGGCGUGGCAUGGGCGUUGGGUGGCUGCAGGUGGUGGAUUCCCACCCCCGCAACCACAAUUAAACGCAC ......(((((((((...........)))))))))....(((((..((((.(((.((.(((((....))))))).))).)))).....))))).. ( -47.40) >DroPse_CAF1 109604 75 + 1 AAUUCGGCCACGCUUAAUGGCCAAAACAGCAUGGCUUGGGGA------GGG-GGUGGGGGGGC-------------AAGCCACAAUUAAAUGCAC ..(((.(((.(.(((...(((((........)))))....))------).)-))).))).((.-------------...)).............. ( -19.10) >DroSec_CAF1 53475 95 + 1 AAUUUGGCCACGCCUAAUGACCAAAAAGGCGUGGCAUGGGCGUUGGGUGGCUGCAGGUGGUGGCUUCCCACCCCCUCAACCACAAUUAAACGCAC ......(((((((((...........)))))))))....(((((..((((.((.(((.(((((....))))).))))).)))).....))))).. ( -44.30) >DroSim_CAF1 55325 95 + 1 AAUUUGGCCACGCCUAAUGACCAAAAAGGCGUGGCAUGGGCGUUGGGUGGCUGCAGGUGGUGGAUUGCCACCCCCUCAACCACAAUUAAACGCAC ......(((((((((...........)))))))))....(((((..((((.((.(((.(((((....))))).))))).)))).....))))).. ( -44.90) >DroYak_CAF1 53927 95 + 1 AAUUCGGCCAUGCCUAAUGACCAAAAAGGCGUGGCAUGGACGUUGGGUGGCUGCAGGUGGUGGAUGACCGCCCCCUCAACCACAAUUAAACGCAC ..(((((((((((((...........))))))))).))))((((..((((.((.(((.(((((....))))).))))).)))).....))))... ( -39.40) >DroPer_CAF1 105683 76 + 1 AAUUCGGCCACGCUUAAUGGCCAAAACAGCAAGGCUUGGGGA------GGGGGGAGGGGGGGC-------------AAGCCACAAUUAAAUGCAC .....(((((.......)))))..........((((((....------..............)-------------))))).............. ( -16.27) >consensus AAUUCGGCCACGCCUAAUGACCAAAAAGGCGUGGCAUGGGCGUUGGGUGGCUGCAGGUGGUGGAUU_CCACCCCCUCAACCACAAUUAAACGCAC ......(((((((((...........)))))))))....(((((..((((.....((.(((((....))))).))....)))).....))))).. (-20.45 = -24.40 + 3.95)

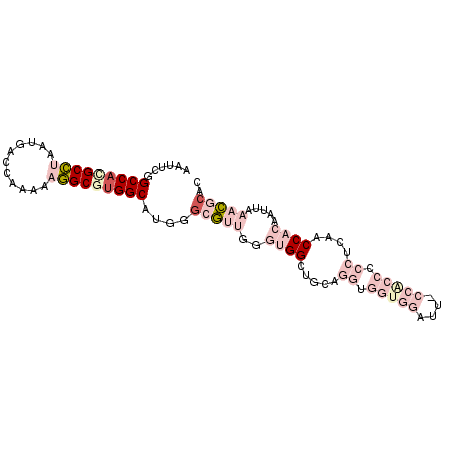
| Location | 17,210,295 – 17,210,390 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -21.04 |
| Energy contribution | -25.52 |
| Covariance contribution | 4.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
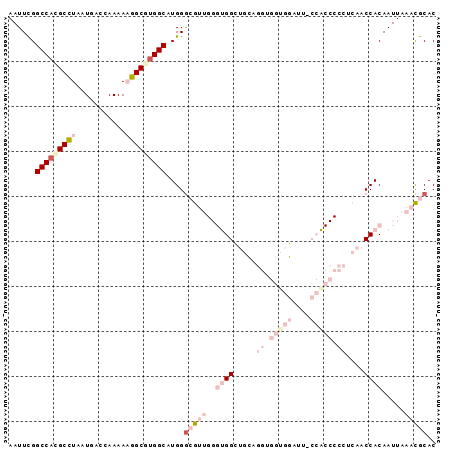
>2L_DroMel_CAF1 17210295 95 - 22407834 GUGCGUUUAAUUGUGGUUGCGGGGGUGGGAAUCCACCACCUGCAGCCACCCAACGCCCAUGCCACGCCUUUUUGGUCAUUAGGCGUGGCCAAAUU (.(((((.....((((((((((((((((....))))).)))))))))))..))))).)..(((((((((...........)))))))))...... ( -51.90) >DroPse_CAF1 109604 75 - 1 GUGCAUUUAAUUGUGGCUU-------------GCCCCCCCACC-CCC------UCCCCAAGCCAUGCUGUUUUGGCCAUUAAGCGUGGCCGAAUU ............(((((((-------------(..........-...------....)))))))).....(((((((((.....))))))))).. ( -21.23) >DroSec_CAF1 53475 95 - 1 GUGCGUUUAAUUGUGGUUGAGGGGGUGGGAAGCCACCACCUGCAGCCACCCAACGCCCAUGCCACGCCUUUUUGGUCAUUAGGCGUGGCCAAAUU (.(((((.....(((((((.((((((((....))))).))).)))))))..))))).)..(((((((((...........)))))))))...... ( -47.70) >DroSim_CAF1 55325 95 - 1 GUGCGUUUAAUUGUGGUUGAGGGGGUGGCAAUCCACCACCUGCAGCCACCCAACGCCCAUGCCACGCCUUUUUGGUCAUUAGGCGUGGCCAAAUU (.(((((.....(((((((.((((((((....))))).))).)))))))..))))).)..(((((((((...........)))))))))...... ( -45.30) >DroYak_CAF1 53927 95 - 1 GUGCGUUUAAUUGUGGUUGAGGGGGCGGUCAUCCACCACCUGCAGCCACCCAACGUCCAUGCCACGCCUUUUUGGUCAUUAGGCAUGGCCGAAUU ............(((((((.(((((.((....)).)).))).)))))))....((.(((((((..(((.....))).....))))))).)).... ( -37.00) >DroPer_CAF1 105683 76 - 1 GUGCAUUUAAUUGUGGCUU-------------GCCCCCCCUCCCCCC------UCCCCAAGCCUUGCUGUUUUGGCCAUUAAGCGUGGCCGAAUU ..(((......)))(((((-------------(..............------....)))))).......(((((((((.....))))))))).. ( -19.17) >consensus GUGCGUUUAAUUGUGGUUGAGGGGGUGG_AAUCCACCACCUGCAGCCACCCAACGCCCAUGCCACGCCUUUUUGGUCAUUAGGCGUGGCCAAAUU (.(((((.....(((((((.(((((.((....)).)).))).)))))))..))))).)..(((((((((...........)))))))))...... (-21.04 = -25.52 + 4.48)


| Location | 17,210,326 – 17,210,420 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -18.89 |
| Energy contribution | -22.23 |
| Covariance contribution | 3.35 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
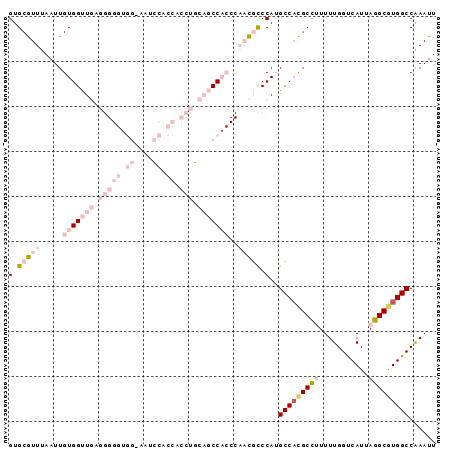
>2L_DroMel_CAF1 17210326 94 + 22407834 U------------GGCAUG---GGCGUUGGGUGGCUGCAGGUGGUGGAUUCCCACCCCCGCAACCACAAUUAAACGCACGCAAAUGGCGAGCCACACACAUAAAACAAA (------------(((...---.(((((..((((.(((.((.(((((....))))))).))).)))).....))))).(((.....))).))))............... ( -38.90) >DroSec_CAF1 53506 94 + 1 U------------GGCAUG---GGCGUUGGGUGGCUGCAGGUGGUGGCUUCCCACCCCCUCAACCACAAUUAAACGCACGCAAAUGGCGGGCCACACACAUAAAACAAA (------------(((...---.(((((..((((.((.(((.(((((....))))).))))).)))).....))))).(((.....))).))))............... ( -35.80) >DroSim_CAF1 55356 94 + 1 U------------GGCAUG---GGCGUUGGGUGGCUGCAGGUGGUGGAUUGCCACCCCCUCAACCACAAUUAAACGCACGCAAAUGGCGGGCCACACACAUAAAACAAA (------------(((...---.(((((..((((.((.(((.(((((....))))).))))).)))).....))))).(((.....))).))))............... ( -36.40) >DroEre_CAF1 53697 92 + 1 UGGGCGUGGGCGUGGCAUG---GGCGUUGGGUGGCUGC--------------CACCCCCUGAACCACAAUUAAACGCACGCAAAUGGCGGGCCACACACAUAAAACAAA ...(.((((((.((.(((.---.((((.((((((...)--------------)))))..((.....)).........))))..))).)).))).))).).......... ( -30.90) >DroYak_CAF1 53958 94 + 1 U------------GGCAUG---GACGUUGGGUGGCUGCAGGUGGUGGAUGACCGCCCCCUCAACCACAAUUAAACGCACGCAAAUGGCGGGCCACACACAUAAAACAAA (------------(((...---(.((((..((((.((.(((.(((((....))))).))))).)))).....))))).(((.....))).))))............... ( -32.00) >DroAna_CAF1 128436 84 + 1 U------------GGCAGCAGAAGCAGGGGCGGGGUGGAGGUGGGGGU-------------GGCCACAAUUAAACGCACGCAAAUUGUGGGGCACACACAUAAAACAAA .------------....((....((((..((((.((....((((....-------------..))))......)).).)))...))))...))................ ( -19.30) >consensus U____________GGCAUG___GGCGUUGGGUGGCUGCAGGUGGUGGAUU_CCACCCCCUCAACCACAAUUAAACGCACGCAAAUGGCGGGCCACACACAUAAAACAAA .............(((.......(((((..((((.((..((.(((((....)))))))..)).)))).....))))).(((.....))).)))................ (-18.89 = -22.23 + 3.35)

| Location | 17,210,326 – 17,210,420 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -17.27 |
| Energy contribution | -20.87 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
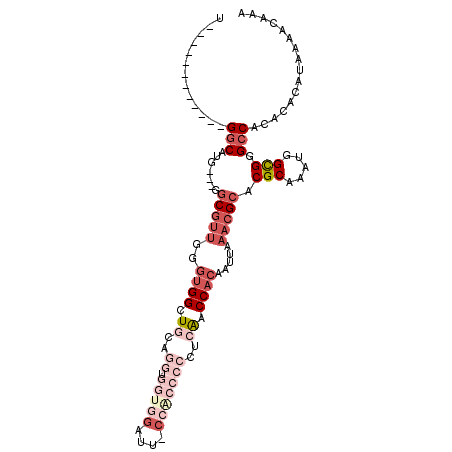
>2L_DroMel_CAF1 17210326 94 - 22407834 UUUGUUUUAUGUGUGUGGCUCGCCAUUUGCGUGCGUUUAAUUGUGGUUGCGGGGGUGGGAAUCCACCACCUGCAGCCACCCAACGCC---CAUGCC------------A .........((.((((((..(((.....))).(((((.....((((((((((((((((....))))).)))))))))))..))))))---))))))------------) ( -42.50) >DroSec_CAF1 53506 94 - 1 UUUGUUUUAUGUGUGUGGCCCGCCAUUUGCGUGCGUUUAAUUGUGGUUGAGGGGGUGGGAAGCCACCACCUGCAGCCACCCAACGCC---CAUGCC------------A .........((.((((((..(((.....))).(((((.....(((((((.((((((((....))))).))).)))))))..))))))---))))))------------) ( -38.50) >DroSim_CAF1 55356 94 - 1 UUUGUUUUAUGUGUGUGGCCCGCCAUUUGCGUGCGUUUAAUUGUGGUUGAGGGGGUGGCAAUCCACCACCUGCAGCCACCCAACGCC---CAUGCC------------A .........((.((((((..(((.....))).(((((.....(((((((.((((((((....))))).))).)))))))..))))))---))))))------------) ( -36.10) >DroEre_CAF1 53697 92 - 1 UUUGUUUUAUGUGUGUGGCCCGCCAUUUGCGUGCGUUUAAUUGUGGUUCAGGGGGUG--------------GCAGCCACCCAACGCC---CAUGCCACGCCCACGCCCA ..........(((((.(((.((((......).))).......(((((...(((((((--------------(...))))))....))---...)))))))))))))... ( -32.00) >DroYak_CAF1 53958 94 - 1 UUUGUUUUAUGUGUGUGGCCCGCCAUUUGCGUGCGUUUAAUUGUGGUUGAGGGGGCGGUCAUCCACCACCUGCAGCCACCCAACGUC---CAUGCC------------A .........((.((((((..(((.....))).(((((.....(((((((.(((((.((....)).)).))).)))))))..))))))---))))))------------) ( -31.10) >DroAna_CAF1 128436 84 - 1 UUUGUUUUAUGUGUGUGCCCCACAAUUUGCGUGCGUUUAAUUGUGGCC-------------ACCCCCACCUCCACCCCGCCCCUGCUUCUGCUGCC------------A ..........(((.(((..((((((((.((....))..)))))))).)-------------))...))).........((....((....)).)).------------. ( -14.70) >consensus UUUGUUUUAUGUGUGUGGCCCGCCAUUUGCGUGCGUUUAAUUGUGGUUGAGGGGGUGG_AAUCCACCACCUGCAGCCACCCAACGCC___CAUGCC____________A ........(((((.((((....)))).)))))(((((.....(((((((.(((((.((....)).)).))).)))))))..)))))....................... (-17.27 = -20.87 + 3.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:23 2006