
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,209,190 – 17,209,329 |
| Length | 139 |
| Max. P | 0.849556 |

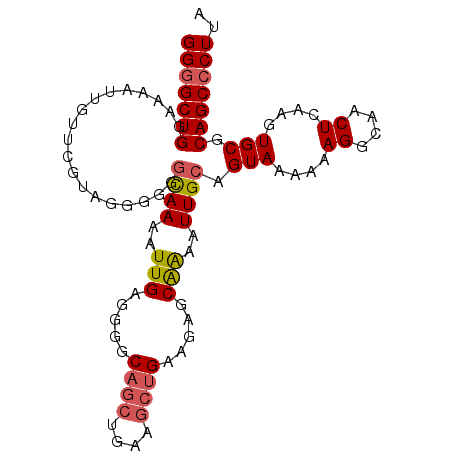
| Location | 17,209,190 – 17,209,289 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17209190 99 + 22407834 GGGGCUGGAAAAUUGUUCGUAGGGGGGCAAAAUUGAGGGGCAGCUGAAGCUGAAGAGCAAAAUUGCAGUAAAAAAGGCAACUCAAGUGCGCAGCCCUUA ((((((((((.....)))........(((...(((((..(((((....))).....(((....)))..........))..))))).))).))))))).. ( -31.60) >DroSec_CAF1 52394 99 + 1 GGGGCUGGUAAAUUGUUCGUAGGGGGGCAAAAUUGAGGGGCAGCUGAAGCUGAAGAGCAAAAUUGCAGUAAAAAAGGCAACUCAAGUGCGCAGCCCUUA ((((((((((..((((((......))))))..(((((..(((((....))).....(((....)))..........))..))))).))).))))))).. ( -33.50) >DroSim_CAF1 54230 99 + 1 GGGGCUGGAAAAUUGUUCGUAGGGGGGCAAAAUUGAGGGGCAGCUGAAGCUGAAGAGCAAAAUUGCAGUAAAAAAGGCAACUCAAGUGCGCAGCCCUUA ((((((((((.....)))........(((...(((((..(((((....))).....(((....)))..........))..))))).))).))))))).. ( -31.60) >DroEre_CAF1 52656 97 + 1 GGGGCUGGAAAAUUGUUCGCAGG-AGGUAAAAUUGAAG-GCAGCUGAAGCUGAAGAGCGAAAUUGCAGUAAAAAAGGCAACUCAAGUGCGCAGCCCUUA ((((((((((.....)))(((.(-((......(((...-.((((....)))).....)))..((((..(....)..)))))))...))).))))))).. ( -27.50) >DroYak_CAF1 52845 91 + 1 GGGGCUGGAAAAUUGUUCGUGGG-AGGUAAAAUUGAAG-GCAGCU------GAAGAGCGAAAUUGCAGUAAAAAAGGCAACUCAAGUGCGCAGCCCUUA (((((((.....((((((.(.((-..((..........-))..))------.).)))))).......(((....((....))....))).))))))).. ( -24.50) >DroAna_CAF1 127196 95 + 1 GGCGCUGGCGAGG-GGGC---GUGGCACAAGAUUGAGCGGCAGCUGAAGCUGAAGAGCGGAAUUGCAGUAAAAAAGGCAGCUGAAGUGCGCAGCCCUUA .((....))..((-((((---.((((((..((((..((..((((....))))....))..)))).((((..........))))..)))).)))))))). ( -30.80) >consensus GGGGCUGGAAAAUUGUUCGUAGGGGGGCAAAAUUGAGGGGCAGCUGAAGCUGAAGAGCAAAAUUGCAGUAAAAAAGGCAACUCAAGUGCGCAGCCCUUA (((((((...................((((..(((.....((((....)))).....)))..)))).(((....((....))....))).))))))).. (-23.76 = -23.98 + 0.22)

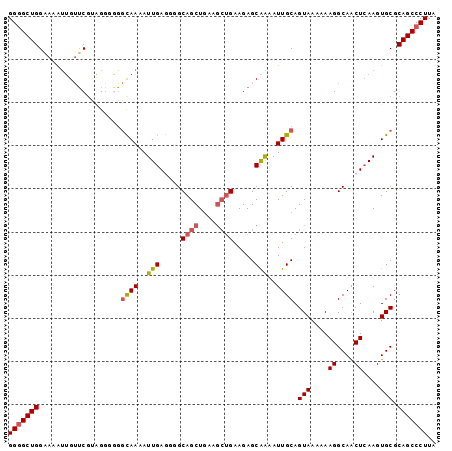
| Location | 17,209,218 – 17,209,329 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17209218 111 - 22407834 GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGGGC-U-GCGCACUUGAGUUGCCUUUUUUACUGCAAUUUUG------CUCUU-CAGCUUCAGCUGCCCCUCAAUUUU (((...(((((((((....))))))..)))..)))........((((-.-..(((...(((((((..........)))))))))------)....-.(((....)))))))......... ( -25.90) >DroVir_CAF1 147674 91 - 1 GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGG-C-A-GCGCACUUGAGUUGCCUUUUUUACUGCAAUUUCG------UCCCUCCGUC-------------------- ((((..(((((((((....))))..................((((-(-(-((........)))))))).......)))))...)------))).......-------------------- ( -21.90) >DroPse_CAF1 107788 112 - 1 GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGGGC-UGGCGCACUUGAGUUGCCUUUUUUACUGCAAUUUCG------CUCUU-CAGACUCAGCAGAUGCCCCAGCCC (((...(((((((((....))))))..)))..)))........((((-(((.(((((((((((............((......)------)....-..))))))..)).))).))))))) ( -33.37) >DroGri_CAF1 171872 102 - 1 GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGG-C-A-GCGCACUUGAGUUGCCUUUUUUACUGCAAUUUCG------UCCUU-CAACUGCUAUUCCCUC-------- ((((..(((((((((....))))..................((((-(-(-((........)))))))).......)))))...)------)))..-................-------- ( -22.30) >DroWil_CAF1 62669 104 - 1 GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGGCUGU-GCGCACUUGAGUUGCCUUUUUUACUGCAAUUUCUGCGUCCUUCUU-CAG-UUCAGUU------------- (((...(((((((((....))))))..)))..)))......((((....-(((((...(((((((..........))))))).)))))))))...-...-.......------------- ( -21.70) >DroMoj_CAF1 119349 102 - 1 GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGG-C-A-GCGCACUUGAGUUGCCGUUUUUACUGCAAUUUCG------UCCUU-CGUCAGCGCCGUCAUG-------- (((...(((((((((....))))))..)))..)))........((-(-.-((((....(((((((.((....)).)))))))((------.....-))...)))).)))...-------- ( -26.90) >consensus GGAUUUGUUGUGCUUAUGAAAGCAUUAAACUUUCCUCAUUUAAGG_C_A_GCGCACUUGAGUUGCCUUUUUUACUGCAAUUUCG______UCCUU_CAGCUUCAGCU_C__C________ (((...(((((((((....))))))..)))..))).......................(((((((..........)))))))...................................... (-13.20 = -13.20 + 0.00)


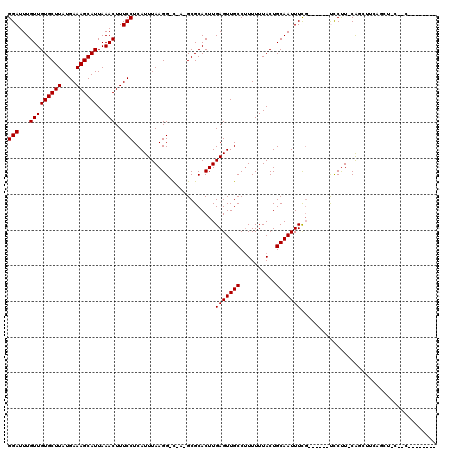
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:19 2006