
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,169,311 – 17,169,452 |
| Length | 141 |
| Max. P | 0.950194 |

| Location | 17,169,311 – 17,169,425 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -9.68 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.922003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17169311 114 + 22407834 UAAGUACUGGAAUCAGACGUCAUAAAGA---AUUUGUGGAUACUAGACUACUAGUUUGACCUUCGAAA-CAAAACAGGCCUUUAUUUGCCUGUCUGUUUUGUAUUUAAUCCUUUUCCA (((((((.(((.((((((.((((((...---..))))))....(((....)))))))))..)))((((-((..((((((........)))))).)))))))))))))........... ( -25.60) >DroSec_CAF1 14790 114 + 1 UAAAGACUUGAAUCAGACUGCAUAAAGA---AUUUGUGGAAACUAGACAUCUAGUUAUACCUUUGAAA-CAGAACAGGCCUUUAUGUGCCUGUCUGUUUUGUUUUUUAUCCUUUUCCA .((((((.....((((((..((......---...))..).((((((....)))))).....)))))((-((((.((((((.....).)))))))))))..))))))............ ( -28.00) >DroSim_CAF1 14768 103 + 1 UAAAG-----------ACUGCAUAAAGA---AUUUGUGGAAACUAGACAUCUAGUUAUACGUUUGAAA-CAAAACAGGCCUUUAUGUGCCUGUCUGUUUUGUUUUUUAUUCUUUUCCA .((((-----------(....(((((((---((..(((..((((((....)))))).)))....((((-((..(((((((.....).)))))).)))))))))))))))))))).... ( -25.60) >DroEre_CAF1 14915 105 + 1 UCAGGACUGGAAUCAGACUUCAUAAAGAAAACUUUGUGAAUACAAGAC------UU-------UGUAAUCUAAGCAGGCCUUUUUGUGCCUGUUCGAUUGGUAUUUUACCAUUUUCCA ...(((....(((((((.(((((((((....)))))))))(((((...------.)-------)))).))).((((((((.....).))))))).))))((((...))))....))). ( -27.10) >DroYak_CAF1 15845 112 + 1 UUAAGACGGAAAUCAGAAAGCAUGAAAAAAAAUUGGUGAAUACUAGAG------UUCUACCUCCGAAAUCAAAACAGGCCUUUUUGUGAAUGUCUGAUUGGUAUUUUAAUAUUUUCCA .......((((((((((...((..((((......(((((((......)------))).))).((............))..))))..))....)))))))(((((....))))).))). ( -19.90) >consensus UAAAGACUGGAAUCAGACUGCAUAAAGA___AUUUGUGGAUACUAGAC__CUAGUUAUACCUUUGAAA_CAAAACAGGCCUUUAUGUGCCUGUCUGUUUUGUAUUUUAUCCUUUUCCA ....................((((((......))))))(((((.((((................(....)...((((((........))))))..)))).)))))............. ( -9.68 = -10.00 + 0.32)

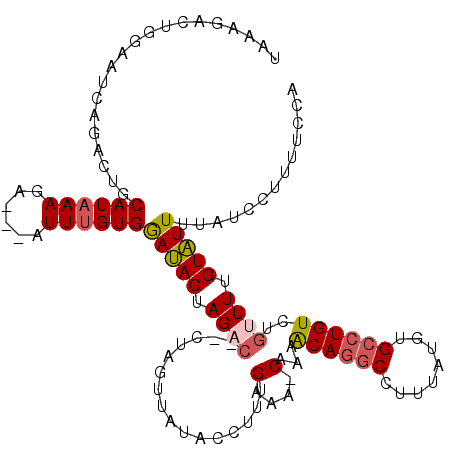

| Location | 17,169,311 – 17,169,425 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -9.44 |
| Energy contribution | -11.32 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17169311 114 - 22407834 UGGAAAAGGAUUAAAUACAAAACAGACAGGCAAAUAAAGGCCUGUUUUG-UUUCGAAGGUCAAACUAGUAGUCUAGUAUCCACAAAU---UCUUUAUGACGUCUGAUUCCAGUACUUA (((((..((((...((((.((((((((((((........))))).))))-))).)..((....(((((....)))))..))......---....)))...))))..)))))....... ( -28.30) >DroSec_CAF1 14790 114 - 1 UGGAAAAGGAUAAAAAACAAAACAGACAGGCACAUAAAGGCCUGUUCUG-UUUCAAAGGUAUAACUAGAUGUCUAGUUUCCACAAAU---UCUUUAUGCAGUCUGAUUCAAGUCUUUA ..(((..((((........((((((((((((........))))).))))-)))....((...((((((....)))))).))......---..........))))..)))......... ( -29.00) >DroSim_CAF1 14768 103 - 1 UGGAAAAGAAUAAAAAACAAAACAGACAGGCACAUAAAGGCCUGUUUUG-UUUCAAACGUAUAACUAGAUGUCUAGUUUCCACAAAU---UCUUUAUGCAGU-----------CUUUA ....(((((((........((((((((((((........))))).))))-))).....((..((((((....))))))...))..))---))))).......-----------..... ( -24.00) >DroEre_CAF1 14915 105 - 1 UGGAAAAUGGUAAAAUACCAAUCGAACAGGCACAAAAAGGCCUGCUUAGAUUACA-------AA------GUCUUGUAUUCACAAAGUUUUCUUUAUGAAGUCUGAUUCCAGUCCUGA (((((..(((((...)))))......(((((........))))).((((((((((-------(.------...)))))((((.((((....)))).)))))))))))))))....... ( -25.00) >DroYak_CAF1 15845 112 - 1 UGGAAAAUAUUAAAAUACCAAUCAGACAUUCACAAAAAGGCCUGUUUUGAUUUCGGAGGUAGAA------CUCUAGUAUUCACCAAUUUUUUUUCAUGCUUUCUGAUUUCCGUCUUAA .(((((.............(((((((((..(........)..)).)))))))((((((((((((------....(((........)))....))).))).)))))))))))....... ( -14.70) >consensus UGGAAAAGGAUAAAAUACAAAACAGACAGGCACAUAAAGGCCUGUUUUG_UUUCAAAGGUAUAACUAG__GUCUAGUAUCCACAAAU___UCUUUAUGCAGUCUGAUUCCAGUCUUUA ((((...................((((((((........))))))))................(((((....))))).)))).................................... ( -9.44 = -11.32 + 1.88)

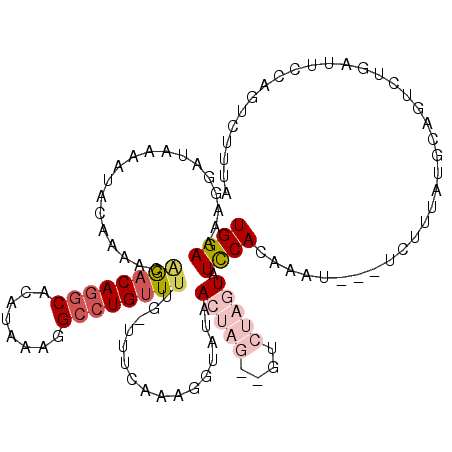
| Location | 17,169,348 – 17,169,452 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -20.71 |
| Consensus MFE | -7.02 |
| Energy contribution | -7.86 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17169348 104 + 22407834 UACUAGACUACUAGUUUGACCUUCGAAA-CAAAACAGGCCUUUAUUUGCCUGUCUGUUUUGUAUUUAAUCCUUUUCCAUAAGCUAGAUCAUAAAAUGU-------------CUUGCUA .(((((....)))))((((....(((((-((..((((((........)))))).)))))))...))))............(((.((((........))-------------)).))). ( -22.30) >DroSec_CAF1 14827 104 + 1 AACUAGACAUCUAGUUAUACCUUUGAAA-CAGAACAGGCCUUUAUGUGCCUGUCUGUUUUGUUUUUUAUCCUUUUCCAUAAGCUAGAGCAUAAAAUGU-------------CUUACUA ((((((....))))))........((((-((((.((((((.....).)))))))))))))...((((((.((((..(....)..)))).))))))...-------------....... ( -26.00) >DroSim_CAF1 14794 104 + 1 AACUAGACAUCUAGUUAUACGUUUGAAA-CAAAACAGGCCUUUAUGUGCCUGUCUGUUUUGUUUUUUAUUCUUUUCCAUAAGCUAGAGCAUAAAAUGU-------------CUUACUA ((((((....))))))..((((((((((-((((((((((............))))))))))))))................((....))...))))))-------------....... ( -23.60) >DroEre_CAF1 14955 105 + 1 UACAAGAC------UU-------UGUAAUCUAAGCAGGCCUUUUUGUGCCUGUUCGAUUGGUAUUUUACCAUUUUCCAUAAGCUAGAAUAUAAAAUGUUCCUCUAAAAAGACUCACUA ........------((-------((((.((((((((((((.....).))))))).((.(((((...)))))...)).......)))).))))))........................ ( -19.10) >DroYak_CAF1 15885 112 + 1 UACUAGAG------UUCUACCUCCGAAAUCAAAACAGGCCUUUUUGUGAAUGUCUGAUUGGUAUUUUAAUAUUUUCCAAAAGCUAGAAUAUAAAAUGUACUUCUAAAAAGUCUUACUA .......(------(((((.((............(((((.(((....))).))))).((((..............)))).)).))))))............................. ( -12.54) >consensus UACUAGAC__CUAGUUAUACCUUUGAAA_CAAAACAGGCCUUUAUGUGCCUGUCUGUUUUGUAUUUUAUCCUUUUCCAUAAGCUAGAACAUAAAAUGU_____________CUUACUA ..((((.......................((((((((((............)))))))))).........(((......)))))))................................ ( -7.02 = -7.86 + 0.84)

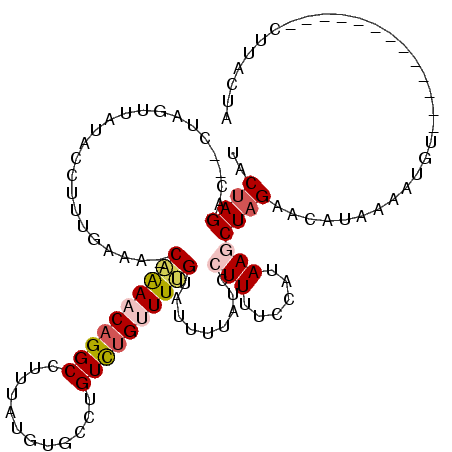
| Location | 17,169,348 – 17,169,452 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17169348 104 - 22407834 UAGCAAG-------------ACAUUUUAUGAUCUAGCUUAUGGAAAAGGAUUAAAUACAAAACAGACAGGCAAAUAAAGGCCUGUUUUG-UUUCGAAGGUCAAACUAGUAGUCUAGUA ..((.((-------------(((((...((((((..(((......))).........(.((((((((((((........))))).))))-))).).))))))....))).)))).)). ( -24.10) >DroSec_CAF1 14827 104 - 1 UAGUAAG-------------ACAUUUUAUGCUCUAGCUUAUGGAAAAGGAUAAAAAACAAAACAGACAGGCACAUAAAGGCCUGUUCUG-UUUCAAAGGUAUAACUAGAUGUCUAGUU .....((-------------(((((((((((((((.....))))...............((((((((((((........))))).))))-))).....))))))...))))))).... ( -27.90) >DroSim_CAF1 14794 104 - 1 UAGUAAG-------------ACAUUUUAUGCUCUAGCUUAUGGAAAAGAAUAAAAAACAAAACAGACAGGCACAUAAAGGCCUGUUUUG-UUUCAAACGUAUAACUAGAUGUCUAGUU .....((-------------(((((((((((((((.....))))...............((((((((((((........))))).))))-))).....))))))...))))))).... ( -25.60) >DroEre_CAF1 14955 105 - 1 UAGUGAGUCUUUUUAGAGGAACAUUUUAUAUUCUAGCUUAUGGAAAAUGGUAAAAUACCAAUCGAACAGGCACAAAAAGGCCUGCUUAGAUUACA-------AA------GUCUUGUA ..(..((.((((.....((...(((((((.(((((.....)))))....))))))).))((((...(((((........)))))....))))..)-------))------).))..). ( -21.60) >DroYak_CAF1 15885 112 - 1 UAGUAAGACUUUUUAGAAGUACAUUUUAUAUUCUAGCUUUUGGAAAAUAUUAAAAUACCAAUCAGACAUUCACAAAAAGGCCUGUUUUGAUUUCGGAGGUAGAA------CUCUAGUA ..(((...((....))...)))........(((((.((((((((....(((((((((((...................))..))))))))))))))))))))))------........ ( -13.41) >consensus UAGUAAG_____________ACAUUUUAUGCUCUAGCUUAUGGAAAAGGAUAAAAUACAAAACAGACAGGCACAUAAAGGCCUGUUUUG_UUUCAAAGGUAUAACUAG__GUCUAGUA ...............................((((.....))))...................((((((((........))))))))............................... ( -9.00 = -9.52 + 0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:08 2006