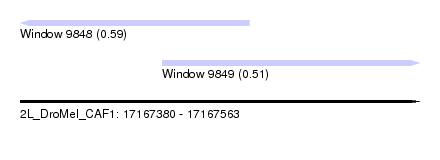
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,167,380 – 17,167,563 |
| Length | 183 |
| Max. P | 0.585249 |

| Location | 17,167,380 – 17,167,485 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.73 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17167380 105 - 22407834 CGCCGUGUCUCCUCGAUUUGGCUGCCCACAGUUCUGUGGUGCAAGAGUUCAGCUGUGAAUAUCACCUGGUCAUACAUCUGACUUAAACU---------------AGGAUGUGUAAGUUCU .((((.(((.....))).))))((((((((....))))).)))((((..(((..(((.....))))))..)((((((((..........---------------.))))))))....))) ( -30.00) >DroSec_CAF1 12925 104 - 1 CGCCGUGUCUCCUCGAUUUGGCUGCCCACAGUUCUGUGGUGCAAGAGUUCACAUGUGAAUAUCACCUGGUCAUACAUCCUAAUUA-ACU---------------GCGAUGUGUACGUUCU .((((.(((.....))).))))((((((((....))))).)))((((....((.(((.....))).))((.((((((((......-...---------------).))))))))).)))) ( -29.10) >DroSim_CAF1 12884 105 - 1 CGCCGUGUCUCCUCGAUUUGGCUGCCCACAGUUCUGUGGUGCAAGAGUUCACAUGUGAAUAUCACCUGGUCAUACAUCCUAAUUAUACU---------------GCGAUGUGUACGUUCU .((((.(((.....))).))))((((((((....))))).)))((((....((.(((.....))).))((.((((((((..........---------------).))))))))).)))) ( -29.00) >DroEre_CAF1 13108 102 - 1 CGCCGUGUCUCCUUGAUUCGGCUGCCCACAGAUCUGUGGUGCAACAGCUCAGCUGUGAAUAUCACCUGGUCAUAUACCUUCAUUACAGUUUUUAACGAACUA------------------ .((((.(((.....))).))))((((((((....))))).)))(((((...)))))...........(((.....)))........(((((.....))))).------------------ ( -27.60) >DroYak_CAF1 13826 115 - 1 CGCCGUGUCUCCUUGAUUUGGUUGCCCACAGAUCUGUGGUGCAAAAGCUGUUCUGUGAAUAUCACCUGGUCAUAAACCUUCAUUAUAUU-----GCAAAAUAUUGGAAUGUGUACGUUCA .((((.(((.....))).))))((((((((....))))).)))...((.((...(((((........(((.....))))))))...)).-----)).........(((((....))))). ( -25.80) >DroAna_CAF1 80409 97 - 1 UCUCGAGUUGCCUGGAUCUGUUGGCCCACACAUCGGUGGUGCAACAGCUGGUCGGUGAAGAUCACCUCCUCGUAACCUUUCAUG-----------------------CUUUAAAUAUUUU ......(((((..(((.((((((..((((......))))..))))))......((((.....)))))))..)))))........-----------------------............. ( -26.90) >consensus CGCCGUGUCUCCUCGAUUUGGCUGCCCACAGAUCUGUGGUGCAAGAGCUCACCUGUGAAUAUCACCUGGUCAUACACCUUAAUUA_ACU_______________G_GAUGUGUACGUUCU .((((.(((.....))).))))((((((((....))))).)))...........(((.....)))....................................................... (-16.72 = -17.00 + 0.28)

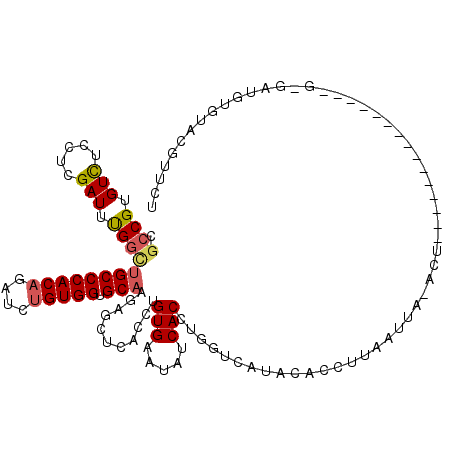
| Location | 17,167,445 – 17,167,563 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -29.05 |
| Energy contribution | -30.47 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17167445 118 + 22407834 ACCACAGAACUGUGGGCAGCCAAAUCGAGGAGACACGGCGCCAUUGGGA--GGAGCGGUGCUCUUGGUUUCCGGCUGCCCAAAGGAAUAUGGCAUCAAGGUGUUCAGAAAUUUACAAGGA .((......((.(((((((((((((((((.((.(((.((.((.......--)).)).)))))))))))))..))))))))).))((((.((((((....))).)))...))))....)). ( -44.80) >DroSec_CAF1 12989 118 + 1 ACCACAGAACUGUGGGCAGCCAAAUCGAGGAGACACGGCGCACUUGGGA--GGAGCGGUGCUCUUGGUUUCCGGCUGCCCAAAGGAAUUUGGCAGCAAGGUGUUCCGAAAUUUACAAGGA .((......((.(((((((((((((((((.((.(((.((...((....)--)..)).)))))))))))))..))))))))).))...(((((.(((.....))))))))........)). ( -45.30) >DroSim_CAF1 12949 118 + 1 ACCACAGAACUGUGGGCAGCCAAAUCGAGGAGACACGGCGCACUUGGGA--GGAGCGGUGCUCUUGGUUUCCGGCUGCCCAAAGGAAUUUGGCAGCAAGGUGUUCCGAAAUUUACAAGGA .((......((.(((((((((((((((((.((.(((.((...((....)--)..)).)))))))))))))..))))))))).))...(((((.(((.....))))))))........)). ( -45.30) >DroEre_CAF1 13170 118 + 1 ACCACAGAUCUGUGGGCAGCCGAAUCAAGGAGACACGGCGCCAUUGGGA--GGAGCGGUGCUCCUGGUUUCCGGCUGCCCAAAGGAAUUUGGAAGCAAGGUGUUCUGAAAUUUACAAGGA ....(((((((.(((((((((((((((.((((.(((.((.((.......--)).)).))))))))))))..)))))))))).)))..((((....))))....))))............. ( -49.70) >DroYak_CAF1 13901 118 + 1 ACCACAGAUCUGUGGGCAACCAAAUCAAGGAGACACGGCGCCAUUGGGA--GGAGCGGUGCUCUUGGUUUCCGGCUGCCCAAAGGUAUUUGGAAGCAAGGUGUUCUGAAAUUUACAAGGA .((....((((.((((((.((((((((((.((.(((.((.((.......--)).)).)))))))))))))..)).)))))).)))).((..(((........)))..))........)). ( -40.30) >DroAna_CAF1 80466 118 + 1 ACCACCGAUGUGUGGGCCAACAGAUCCAGGCAACUCGAGAAGCCUGGGAAAGAAACUGUG--CGUGGUUUCCAGUUGCUCAAAGGAAAAUGUAUGCCAGGUACCCGGAGAUCUAUGAAGA ..((..(((.(.((((........(((((((..(....)..))))))).......(((.(--((((.(((((...........)))))...))))))))...)))).).)))..)).... ( -34.30) >consensus ACCACAGAACUGUGGGCAGCCAAAUCGAGGAGACACGGCGCCAUUGGGA__GGAGCGGUGCUCUUGGUUUCCGGCUGCCCAAAGGAAUUUGGAAGCAAGGUGUUCCGAAAUUUACAAGGA .(((.....((.((((((((((((((.(((((.(((.((.(...........).)).)))))))))))))..))))))))).)).....)))............................ (-29.05 = -30.47 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:05 2006