| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,161,396 – 17,161,504 |
| Length | 108 |
| Max. P | 0.636530 |

| Location | 17,161,396 – 17,161,504 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -14.11 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
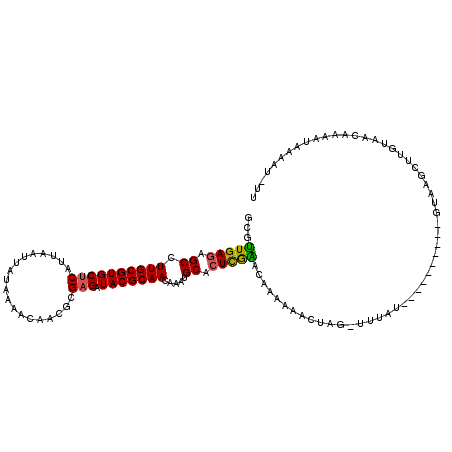
>2L_DroMel_CAF1 17161396 108 + 22407834 GCGUUGAGAGCCUUGCGUGCUCAUUAAUUAUAAAACAACGCGAGAUACGCAACAAAUGCACUCGGACAAGAAACUCG-UUUAU----------UUAAGCUUGUACCAAAAUAAAAUUUA ((((((.(((((....).)))).(((....)))..))))))(((....(((.....))).)))(((((((.......-.....----------.....))))).))............. ( -21.13) >DroSec_CAF1 6802 108 + 1 GCGUUGAAAGCCUUGCGUGCUCAUUAAUUAUAAAACAACGCGAGAUACGCAACAAAUGCACUCGAACAAGAAACUAG-UUUAU----------GUAAGCUUGUACCAAAAUAAAAUGUU (((((....(((((((((...................)))))))....))....)))))......(((((..((...-.....----------))...)))))................ ( -18.21) >DroSim_CAF1 6849 107 + 1 GCGUUGAGAGCCUUGCGUGCUCAUUAAUUAUAAAACAACGCGAGAUACGCAACAAAUGCACUCGAACAAGAAACUCG-UUUAU----------GAAAACUUGUAACAAAA-AAAAUGUU ((((((((.((.((((((((((...................))).))))))).....)).)))..(((((....(((-....)----------))...))))).......-..))))). ( -18.61) >DroEre_CAF1 7389 96 + 1 GCGUUGAGAGCCUUGCGUGCUCAUUAAUUAUAAAACAACGCGAGAUACGCAACAAAUGCACUCAAACAAAU---------UAU----------UUUAGCUAGUAGUAAAUAA-AAG--- ...(((((.((.((((((((((...................))).))))))).....)).)))))....((---------(((----------(......))))))......-...--- ( -17.01) >DroYak_CAF1 7516 105 + 1 GCGUUGAGAGCCUUGCGUGCUCAUUAAUUAUAAAACAACGCGAGAUACGCAACAAAUGCACUUAACCAAAAUUUUAUCCCUAU----------UUUAGCUGGUGAUAAAAUA-AUA--- (.((((((.((.((((((((((...................))).))))))).....)).)))))))...((((((((((...----------.......)).)))))))).-...--- ( -23.91) >DroAna_CAF1 74115 116 + 1 GUGCUGAGAGCCUUGCGUGCUCAUUAAUUAUAAAAAAAAACGCGAUACGCAACAGCUGCACUUGGAGAAAACUUUUA-AUUGUGUUGCCCAUUGUGUGCUUUCAAC--UACAAAGCCCU (((.(((((((.((((((.....................)))))).((((((..((.((((...((((....)))).-...)))).))...)))))))))))))..--)))........ ( -26.40) >consensus GCGUUGAGAGCCUUGCGUGCUCAUUAAUUAUAAAACAACGCGAGAUACGCAACAAAUGCACUCGAACAAAAAACUAG_UUUAU__________GUAAGCUUGUAACAAAAUAAAAU_UU ...(((((.((.((((((((((...................))).))))))).....)).)))))...................................................... (-14.11 = -13.64 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:03 2006