| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,829,125 – 1,829,230 |
| Length | 105 |
| Max. P | 0.626849 |

| Location | 1,829,125 – 1,829,230 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -5.27 |
| Energy contribution | -6.51 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1829125 105 + 22407834 UGCCAAAUUUAAGCCUCUUACUGCGGAAAAUAGAAGAAAAGGCUGGAAAACAUAGGUAGCUGU-----------UUUUACCAGAGGUUCUUACCUAU----AUCUUUCAUUAAAAUGGAA ..(((......((((((((.(((.......)))))))...)))).((((..((((((((..(.-----------.((.....))..)..))))))))----...)))).......))).. ( -19.50) >DroSec_CAF1 49158 104 + 1 UGCCAAAUUUAAGCCCCUUUCUACGGAAAAUAGAAGAAAAGGCUGGAAAACAUAGGUACCUCC------------UUUGCAAAGCGUUCUUGCCUAU----AUCUUCUAUUAAAAUGAAA ..................(((....)))(((((((((..((((.((((..((.(((.....))------------).)).......)))).))))..----.)))))))))......... ( -23.00) >DroSim_CAF1 29721 102 + 1 UGCCAAAUUUAAGCCUCUUUAUACGGAAAAUAGAGGAAAAGUCUGGAAAACAUAAGUAGCUC-------------UUUGCAAAGAGUUCUUGCCUAU----AUCUUCUAUUAA-AUGGAA ..(((.((((...(((((.............)))))..)))).)))......((((.(((((-------------((....))))))))))).....----....(((((...-))))). ( -19.02) >DroEre_CAF1 37720 85 + 1 UGCCAAAUUUAAGCCU----CUACGGCAAAUAGAAGAAAAGGUUGGAAAACAUAUGUUGUU--------------------------CCUUACCGAU----GUCUUUCAUUGA-AUGGCA .((((.......(((.----....))).(((..((((...(((.(((((((....))).))--------------------------))..)))...----.))))..)))..-.)))). ( -24.10) >DroYak_CAF1 14125 119 + 1 UGCCAAAUUUAAGCCUCUUUCUACGAAAAAUAGAAGAAUAGGUUGGAAAACAUAUGUAGUCUUCAUAGAUACAGCUUUGCUGGAUUUUUUAACCUACGUUUUUUUUUUAUUAA-AUGGCA .((((............((((...))))((((((((((((((((((((((........((((....)))).((((...))))..)))))))))))).....))))))))))..-.)))). ( -26.10) >consensus UGCCAAAUUUAAGCCUCUUUCUACGGAAAAUAGAAGAAAAGGCUGGAAAACAUAGGUAGCUC_____________UUUGCAAAA_GUUCUUACCUAU____AUCUUUUAUUAA_AUGGAA ............................(((((((((..((((.((((.......((((.................))))......)))).)))).......)))))))))......... ( -5.27 = -6.51 + 1.24)

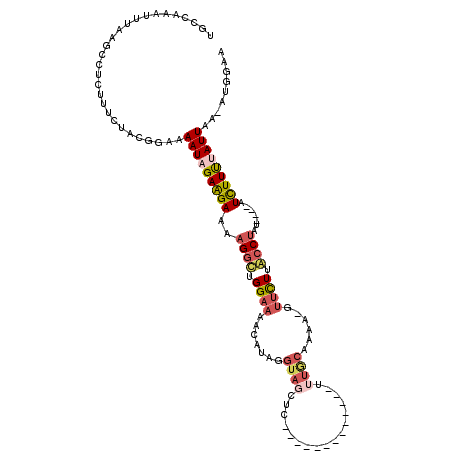
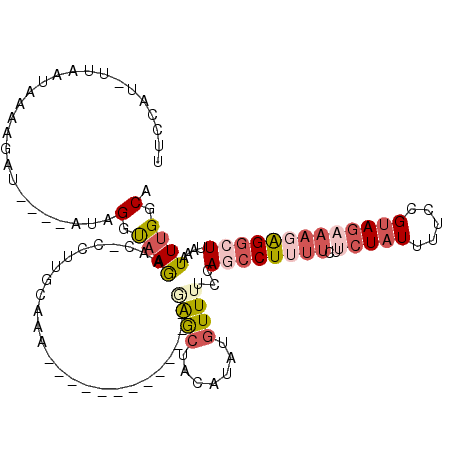
| Location | 1,829,125 – 1,829,230 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.71 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -3.16 |
| Energy contribution | -5.12 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1829125 105 - 22407834 UUCCAUUUUAAUGAAAGAU----AUAGGUAAGAACCUCUGGUAAAA-----------ACAGCUACCUAUGUUUUCCAGCCUUUUCUUCUAUUUUCCGCAGUAAGAGGCUUAAAUUUGGCA ..(((.(((((((.(((((----(((((((.(.....(((......-----------.)))))))))))))))).))(((((((((.(........).)).))))))))))))..))).. ( -24.30) >DroSec_CAF1 49158 104 - 1 UUUCAUUUUAAUAGAAGAU----AUAGGCAAGAACGCUUUGCAAA------------GGAGGUACCUAUGUUUUCCAGCCUUUUCUUCUAUUUUCCGUAGAAAGGGGCUUAAAUUUGGCA .........(((((((((.----..((((......((...))...------------(((((.((....))))))).))))..))))))))).(((.......)))(((.......))). ( -23.80) >DroSim_CAF1 29721 102 - 1 UUCCAU-UUAAUAGAAGAU----AUAGGCAAGAACUCUUUGCAAA-------------GAGCUACUUAUGUUUUCCAGACUUUUCCUCUAUUUUCCGUAUAAAGAGGCUUAAAUUUGGCA ......-......((((((----(((((..((..(((((....))-------------))))).)))))))))))((((.((..(((((.............)))))...)).))))... ( -18.42) >DroEre_CAF1 37720 85 - 1 UGCCAU-UCAAUGAAAGAC----AUCGGUAAGG--------------------------AACAACAUAUGUUUUCCAACCUUUUCUUCUAUUUGCCGUAG----AGGCUUAAAUUUGGCA .((((.-..((((.((((.----...(((..((--------------------------((.(((....))))))).)))...)))).)))).(((....----.))).......)))). ( -19.20) >DroYak_CAF1 14125 119 - 1 UGCCAU-UUAAUAAAAAAAAAACGUAGGUUAAAAAAUCCAGCAAAGCUGUAUCUAUGAAGACUACAUAUGUUUUCCAACCUAUUCUUCUAUUUUUCGUAGAAAGAGGCUUAAAUUUGGCA .(((((-((((............(((((((........((((...)))).......((((((.......)))))).)))))))((((((((.....)))).))))...)))))..)))). ( -24.20) >consensus UUCCAU_UUAAUAAAAGAU____AUAGGUAAGAAC_CCUUGCAAA_____________GAGCUACAUAUGUUUUCCAGCCUUUUCUUCUAUUUUCCGUAGAAAGAGGCUUAAAUUUGGCA ..........................(.((((..........................((((.......))))...((((((((..(((((.....)))))))))))))....)))).). ( -3.16 = -5.12 + 1.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:05 2006