
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,154,179 – 17,154,373 |
| Length | 194 |
| Max. P | 0.972208 |

| Location | 17,154,179 – 17,154,299 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17154179 120 + 22407834 CCCGGGAUCUAGUAGCCGGAGUCCAGCAGGGCAAGUACAAAUUCAUAUGAGAGGGCUGCAAAAGGUUUUUUGUGUUUGCCUUCUGGCCAUUUUGGUGGAAAAAUAUGGCCAGCUGGAAUC .((((..........))))..(((((((((((((((((((((((....))).(((((......))))))))))))))))))).(((((((.((........)).)))))))))))))... ( -42.40) >DroSec_CAF1 115041 120 + 1 CCCGGGAUCUAGUAGCCGGAGUCCAGCAGGACAAGUACAAAUUCAUAUGACAGGGCUGCAAAAGGUUUUUUGUGUUUGCCUUCUGGCCAUUUUGGUUGAAAAAUAUGGCCAGCUGGAAUC .((((......((((((...((((....))))..((.((........))))..))))))..(((((...........)))))((((((((.((........)).)))))))))))).... ( -37.20) >DroSim_CAF1 127229 120 + 1 CCCGGGAUCUAGUAGCCGGAGUCCAGCAGGACAAGUACAAAUUCAUAUGACAGGGCUGCAAAAGGUUUUUUGUGUUUGCCUUCUGGCCAUUUUGGUGGAAAAAUAUGGCCAGCUGGAAUC .((((......((((((...((((....))))..((.((........))))..))))))..(((((...........)))))((((((((.((........)).)))))))))))).... ( -37.20) >DroYak_CAF1 115079 120 + 1 CCCGGGAUCUAGUAGCCAGAGUCCAGCAGGACAAGUACAAAUUCAUGUGACAGGACCGCAAAAGGUUUUUUGUGUUUGGCUUCUGGCCAUUUUGGUGGGAAAAUAUGGCCAGCUGGAAUC .((((........(((((((((((....))))...((((((.((....))..(((((......)))))))))))))))))).((((((((.((........)).)))))))))))).... ( -38.30) >consensus CCCGGGAUCUAGUAGCCGGAGUCCAGCAGGACAAGUACAAAUUCAUAUGACAGGGCUGCAAAAGGUUUUUUGUGUUUGCCUUCUGGCCAUUUUGGUGGAAAAAUAUGGCCAGCUGGAAUC .((((..........))))..(((((((((.((((((((((.((....))..(((((......))))))))))))))))))..(((((((.((........)).)))))))))))))... (-34.62 = -34.75 + 0.12)



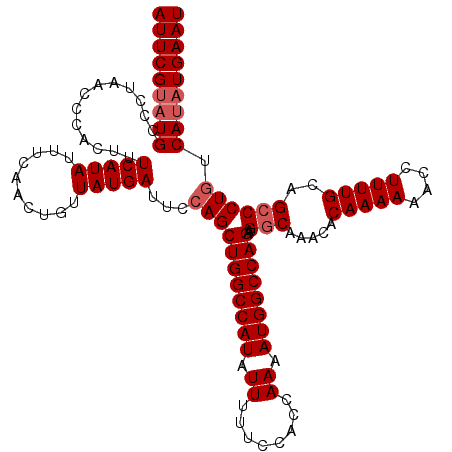
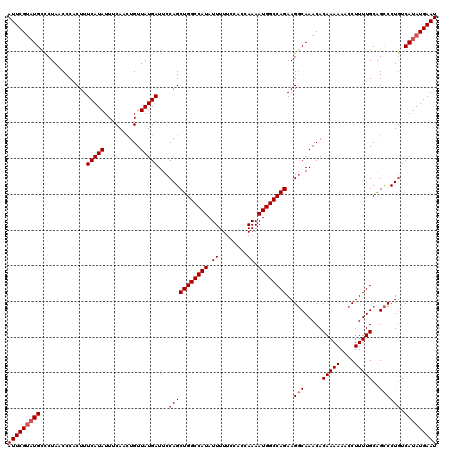
| Location | 17,154,219 – 17,154,339 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -22.02 |
| Energy contribution | -23.02 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17154219 120 - 22407834 AUUCGUAUGCCCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCCACCAAAAUGGCCAGAAGGCAAACACAAAAAACCUUUUGCAGCCCUCUCAUAUGAAU (((((((((.............(((((..........)))))......((((((((.((........)).))))))))..(((.....(((((....)))))..)))....))))))))) ( -24.60) >DroSec_CAF1 115081 120 - 1 AUUCGUAUGUCCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCAACCAAAAUGGCCAGAAGGCAAACACAAAAAACCUUUUGCAGCCCUGUCAUAUGAAU (((((((((.............(((((..........)))))...(((((((((((.((........)).))))))))..(((.....(((((....)))))..)))))).))))))))) ( -26.40) >DroSim_CAF1 127269 120 - 1 AUUCGUAUGCUCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCCACCAAAAUGGCCAGAAGGCAAACACAAAAAACCUUUUGCAGCCCUGUCAUAUGAAU (((((((((.............(((((..........)))))...(((((((((((.((........)).))))))))..(((.....(((((....)))))..)))))).))))))))) ( -26.20) >DroYak_CAF1 115119 120 - 1 AUUCGAAUGCCCUAGCCCGCUUUCAUAUUUCAAUUGUUAUGAUUCCAGCUGGCCAUAUUUUCCCACCAAAAUGGCCAGAAGCCAAACACAAAAAACCUUUUGCGGUCCUGUCACAUGAAU (((((..((...(((.((((..(((((..........)))))......((((((((.((........)).)))))))).......................))))..))).))..))))) ( -22.60) >consensus AUUCGUAUGCCCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCCACCAAAAUGGCCAGAAGGCAAACACAAAAAACCUUUUGCAGCCCUGUCAUAUGAAU (((((((((.............(((((..........)))))...(((((((((((.((........)).))))))))..(((.....(((((....)))))..)))))).))))))))) (-22.02 = -23.02 + 1.00)

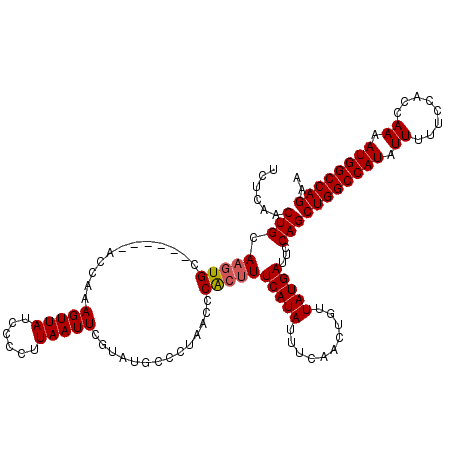
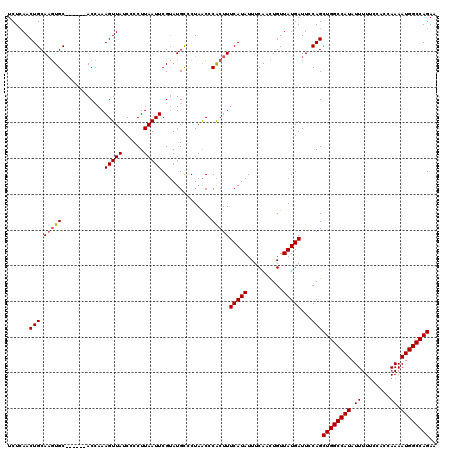
| Location | 17,154,259 – 17,154,373 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.79 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17154259 114 - 22407834 UCUCAACUGCAAGUGC------ACCAAAGUUAUCCCCUUAAUUCGUAUGCCCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCCACCAAAAUGGCCAGAA ......(((..((.((------(....(((((......)))))....))).)).........(((((..........)))))...)))((((((((.((........)).)))))))).. ( -19.20) >DroSec_CAF1 115121 114 - 1 UCUCAACUGCAAGUGC------ACCAAAGUUAUCCCCUUAAUUCGUAUGUCCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCAACCAAAAUGGCCAGAA ......(((..((..(------(....(((((......)))))....))..)).........(((((..........)))))...)))((((((((.((........)).)))))))).. ( -18.20) >DroSim_CAF1 127309 114 - 1 UCUCAACUGCAAGUGC------ACCAAAGUUAUCCCCUUAAUUCGUAUGCUCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCCACCAAAAUGGCCAGAA ......(((..((.((------(....(((((......)))))....))).)).........(((((..........)))))...)))((((((((.((........)).)))))))).. ( -20.00) >DroYak_CAF1 115159 120 - 1 UCUAAACUGCACUUGCACUUACACAAAAGUUAUCCCCUUAAUUCGAAUGCCCUAGCCCGCUUUCAUAUUUCAAUUGUUAUGAUUCCAGCUGGCCAUAUUUUCCCACCAAAAUGGCCAGAA .......(((....)))...........(..(((....(((((.(((((....((....))....))))).)))))....)))..)..((((((((.((........)).)))))))).. ( -19.70) >consensus UCUCAACUGCAAGUGC______ACCAAAGUUAUCCCCUUAAUUCGUAUGCCCUAACCCACUUUCAUAUUUCAACUGUUAUGAUUCCAGCUGGCCAUAUUUUUCCACCAAAAUGGCCAGAA ......(((.(((((............(((((......)))))..............)))))(((((..........)))))...)))((((((((.((........)).)))))))).. (-15.48 = -15.79 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:00 2006