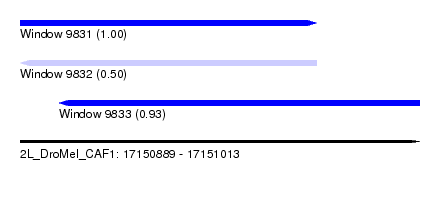
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,150,889 – 17,151,013 |
| Length | 124 |
| Max. P | 0.997949 |

| Location | 17,150,889 – 17,150,981 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
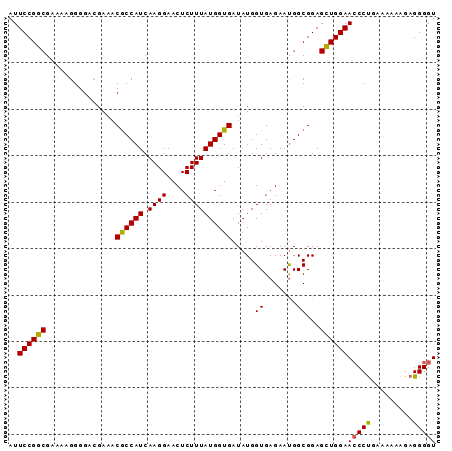
>2L_DroMel_CAF1 17150889 92 + 22407834 AUUCCGGCGAAAAGGGGACGAAACGCCAUCAAGGAACUCUUUAUGGUGAUAUGGUGAGAAUGGCGGAGUUGGAACCCUGAAAAAAGAGGGGU .(((((((.....(....)....((((((.......(((.(((((....))))).))).))))))..)))))))((((........)))).. ( -26.30) >DroSec_CAF1 111772 92 + 1 AUUCCGGCGAAAAGGGGACGAAACGCCAUCAAGGAACUCUUUAUGGUGAUAUGGUGAGAAUGGCGGAGCUGGAACCCUGAAAAAAGAGGGGU .(((((((.....(....)....((((((.......(((.(((((....))))).))).))))))..)))))))((((........)))).. ( -28.80) >DroSim_CAF1 123968 92 + 1 AUUCCGGCGAAAAGGGGACGAAACGCCAUCAAGGAACUCUUUAUGGUGAUAUGGUGAGAAUGGCGGAGCUGGAACCCUGAAAAAAGAGGGGU .(((((((.....(....)....((((((.......(((.(((((....))))).))).))))))..)))))))((((........)))).. ( -28.80) >DroEre_CAF1 109186 90 + 1 AUUCCGGCGAA-AGGGGAAGA-ACACCAUCAAGGAACUCUUUAUGGUGAUAUGGUGAGAAUGGCGGAGCUGGAAGCCUGAAAAAAGAGGGGU .(((((((...-.........-.((((((.((((....)))))))))).....((.......))...))))))).(((........)))... ( -22.00) >DroYak_CAF1 111739 92 + 1 AUUCCGGCGAAAAGGGGAAGAAACGCCAUCAAGGAACUCUUUAUGGUGAUAUGGUGAGAACGGCGGAGCUGGAACCCCGAAAAAAGAGGGGU ..((((((...............((((((.((((....))))))))))...((.((....)).))..))))))(((((.........))))) ( -27.60) >consensus AUUCCGGCGAAAAGGGGACGAAACGCCAUCAAGGAACUCUUUAUGGUGAUAUGGUGAGAAUGGCGGAGCUGGAACCCUGAAAAAAGAGGGGU ..((((((...............((((((.((((....)))))))))).....((.......))...))))))(((((.........))))) (-22.98 = -22.70 + -0.28)

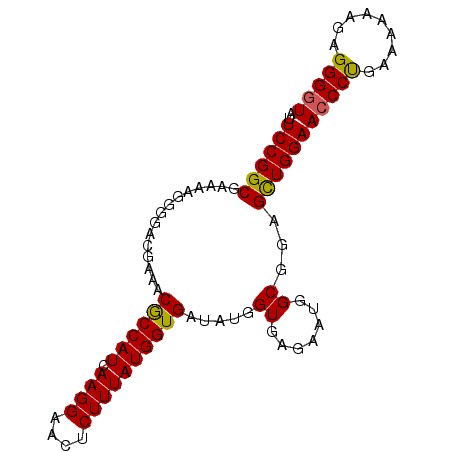
| Location | 17,150,889 – 17,150,981 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -13.79 |
| Energy contribution | -13.51 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17150889 92 - 22407834 ACCCCUCUUUUUUCAGGGUUCCAACUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCUUUUCGCCGGAAU ..((((........))))((((.....((((((((((................)))......)))))))...((.........))..)))). ( -14.99) >DroSec_CAF1 111772 92 - 1 ACCCCUCUUUUUUCAGGGUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCUUUUCGCCGGAAU ..((((........))))((((.((..((((((((((................)))......)))))))....(.........))).)))). ( -16.59) >DroSim_CAF1 123968 92 - 1 ACCCCUCUUUUUUCAGGGUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCUUUUCGCCGGAAU ..((((........))))((((.((..((((((((((................)))......)))))))....(.........))).)))). ( -16.59) >DroEre_CAF1 109186 90 - 1 ACCCCUCUUUUUUCAGGCUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGUGU-UCUUCCCCU-UUCGCCGGAAU ................((.....))((((...............((((((.(((.....))).)))))).-.........-.....)))).. ( -14.21) >DroYak_CAF1 111739 92 - 1 ACCCCUCUUUUUUCGGGGUUCCAGCUCCGCCGUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCUUCCCCUUUUCGCCGGAAU (((((.........)))))......((((.((............(.((((.(((.....))).)))).)..............)).)))).. ( -18.17) >consensus ACCCCUCUUUUUUCAGGGUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCUUUUCGCCGGAAU ...((.......((((((....(((((..........................))))))))))).((((..............))))))... (-13.79 = -13.51 + -0.28)

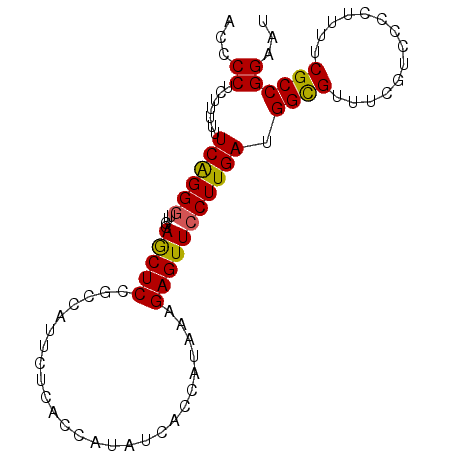
| Location | 17,150,901 – 17,151,013 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.58 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
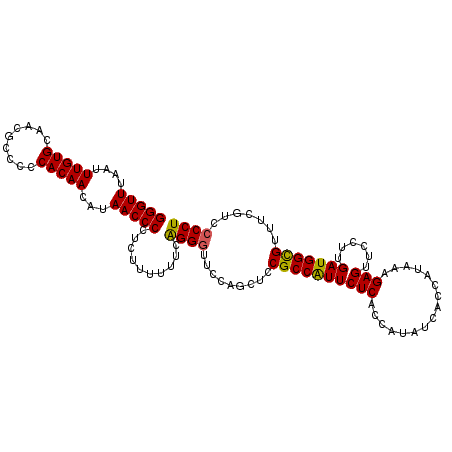
>2L_DroMel_CAF1 17150901 112 - 22407834 GGGUUUAAUUUGUGCAACGCCCCCACAACAUAACCCCUCUUUUUUCAGGGUUCCAACUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCU (((((....(((((.........)))))...)))))..........((((..(......((((((((((................)))......)))))))....)..)))) ( -22.09) >DroSec_CAF1 111784 112 - 1 GGGUUUAAUUUGUGCAACGCCCCCACAACAUAACCCCUCUUUUUUCAGGGUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCU (((((....(((((.........)))))...)))))..........((((..(......((((((((((................)))......)))))))....)..)))) ( -22.09) >DroSim_CAF1 123980 112 - 1 GGGUUUAAUUUGUGCAACGCCCCCACAACAUAACCCCUCUUUUUUCAGGGUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCU (((((....(((((.........)))))...)))))..........((((..(......((((((((((................)))......)))))))....)..)))) ( -22.09) >DroEre_CAF1 109197 111 - 1 GGGUUUAAUUUGUGCAACGCCCCCACAACAUAACCCCUCUUUUUUCAGGCUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGUGU-UCUUCCCCU (((((....(((((.........)))))...)))))...........(((..........))).............((((((.(((.....))).)))))).-......... ( -20.90) >DroYak_CAF1 111751 112 - 1 GGGUUUAAUUUGUGCAACGCCCCCACAACAUAACCCCUCUUUUUUCGGGGUUCCAGCUCCGCCGUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCUUCCCCU (((((....(((((.........)))))...)))))..........((((....((...((((((((((................)))......)))))))...)).)))). ( -24.19) >consensus GGGUUUAAUUUGUGCAACGCCCCCACAACAUAACCCCUCUUUUUUCAGGGUUCCAGCUCCGCCAUUCUCACCAUAUCACCAUAAAGAGUUCCUUGAUGGCGUUUCGUCCCCU (((((....(((((.........)))))...)))))..........((((.........((((((((((................)))......))))))).......)))) (-19.86 = -19.58 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:49 2006