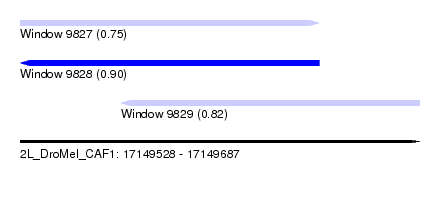
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,149,528 – 17,149,687 |
| Length | 159 |
| Max. P | 0.901067 |

| Location | 17,149,528 – 17,149,647 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -43.86 |
| Consensus MFE | -32.29 |
| Energy contribution | -33.26 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17149528 119 + 22407834 UCUUGCGGUUGCUUUUCCGCCCCGGAAAAUCUCCCACGGGAAAG-GGGCCGGGGCACCCUGACCCCAAGCCAAGCGCGGAAAUUGUGCGCCGCAAUCUCUUAAGACCGGCGACAAAUAUA ..(((((((.((.(((((((((((............))))...(-(((.((((....)))).)))).........)))))))....)))))))))............(....)....... ( -44.90) >DroSec_CAF1 110356 118 + 1 UCUUGCGGUUGCUUUUCCGCCUCGGACAAUCUCUC--GGGAAAGGGGGCAGGGGCCACCUAACCCCAAGCCAAGCGCGGAAAUUGUGCGCCGCAAUCUCUUAAGACCGGCGACAAAUAUA ..(((((((.((.(((((((((.((.(..(((...--.)))..((((..(((.....)))..))))..))).)).)))))))....)))))))))............(....)....... ( -42.60) >DroSim_CAF1 122556 118 + 1 UCUUGCGGUUGCUUUUCCGCCCCGGAAAAUCUCCC--GGGAAAGGGGGCAGGGGCCACCUGACCCCAAGCCAAGCGCGGAAAUUGUGCGCCGCAAUCUCUUAAGACCGGCGACAAAUAUA ..(((((((.((.((((((((((((........))--)))...((((.((((.....)))).)))).........)))))))....)))))))))............(....)....... ( -48.60) >DroEre_CAF1 107828 106 + 1 UCUUGCGGUUGCUUUUCCGC--CGGAAAAUCUCGC--GGGAAAGCGGGC---------CUGACCCCA-ACCAAGCGCGGAAAUUGUGCGCCGCAAUCUCUUAAGACCGGCGACAAAUAUA (((.(((.((((((((((((--.((.....)).))--))))))))(((.---------.....))).-...)).))))))..((((.(((((...(((....))).)))))))))..... ( -39.90) >DroYak_CAF1 110318 118 + 1 UCUUGCGGUUGCUUUUCCGCCCCGGAAAAUCUCCC--GGGAAGGCGGGCAGGGGCCACAUGACCCCAAACCAAGCGCGGAAAUUGUGCGCCGCAAUCUCUUAAGACCGGCGACAAAUAUA ..(((((((.((.((((((((((((........))--)))...((((...((((.(....).))))...))..)))))))))....)))))))))............(....)....... ( -43.30) >consensus UCUUGCGGUUGCUUUUCCGCCCCGGAAAAUCUCCC__GGGAAAGCGGGCAGGGGCCACCUGACCCCAAGCCAAGCGCGGAAAUUGUGCGCCGCAAUCUCUUAAGACCGGCGACAAAUAUA (((.(((.((((((((((.....(((.....)))...))))))).(((((((.....)))).))).....))).))))))..((((.(((((...(((....))).)))))))))..... (-32.29 = -33.26 + 0.97)

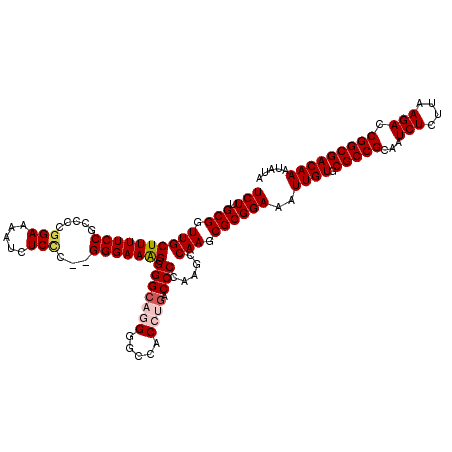
| Location | 17,149,528 – 17,149,647 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -47.98 |
| Consensus MFE | -37.07 |
| Energy contribution | -38.64 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17149528 119 - 22407834 UAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUCAGGGUGCCCCGGCCC-CUUUCCCGUGGGAGAUUUUCCGGGGCGGAAAAGCAACCGCAAGA .....(((((((((((((....))))..))))).))))(((((((.(((((...((((((.((....)).)))))-)(((((....)))))....)))))))))))).((....)).... ( -51.30) >DroSec_CAF1 110356 118 - 1 UAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUUAGGUGGCCCCUGCCCCCUUUCCC--GAGAGAUUGUCCGAGGCGGAAAAGCAACCGCAAGA .....(((((((((((((....))))..))))).))))(((((((.((((((((((((..(((.(((....))).))).))))--))).......)))))))))))).((....)).... ( -48.21) >DroSim_CAF1 122556 118 - 1 UAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUCAGGUGGCCCCUGCCCCCUUUCCC--GGGAGAUUUUCCGGGGCGGAAAAGCAACCGCAAGA .....(((((((((((((....))))..))))).))))(((((((....(((..((((((....)))))).)))......(((--((((....)))))))))))))).((....)).... ( -52.40) >DroEre_CAF1 107828 106 - 1 UAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGU-UGGGGUCAG---------GCCCGCUUUCCC--GCGAGAUUUUCCG--GCGGAAAAGCAACCGCAAGA .....(((((((((((((....))))..))))).)))).......((..(((-(.(((....---------.)))(((((.((--((..(.....)..--)))).))))))))))).... ( -35.80) >DroYak_CAF1 110318 118 - 1 UAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGUUUGGGGUCAUGUGGCCCCUGCCCGCCUUCCC--GGGAGAUUUUCCGGGGCGGAAAAGCAACCGCAAGA .....(((((((((((((....))))..))))).))))(((((((((..(((..((((((....)))))).))).))...(((--((((....)))))))))))))).((....)).... ( -52.20) >consensus UAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUCAGGUGGCCCCUGCCCCCUUUCCC__GGGAGAUUUUCCGGGGCGGAAAAGCAACCGCAAGA .....(((((((((((((....))))..))))).))))(((((((.(((((....(((.((((.....)))))))..(((((....)))))....)))))))))))).((....)).... (-37.07 = -38.64 + 1.57)

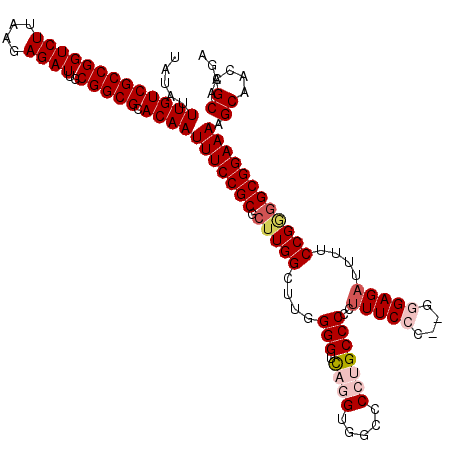
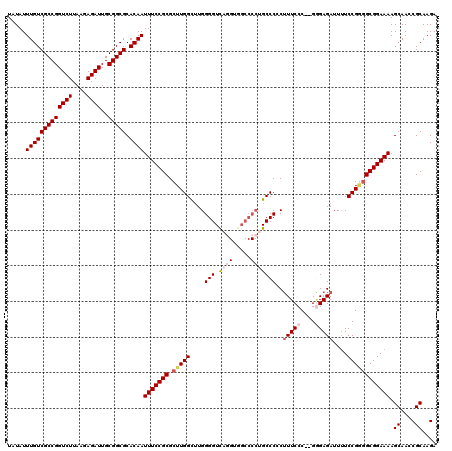
| Location | 17,149,568 – 17,149,687 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -34.54 |
| Energy contribution | -35.34 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17149568 119 - 22407834 CCGCCCCAAAAUCCCAAACCUCCUCCGAUGCCAGGCCAGGUAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUCAGGGUGCCCCGGCCC-CUUU ..(((((((.(((.............)))(((((((..((.....(((((((((((((....))))..))))).))))...))..)))))))))))))).((((.((....))))-)).. ( -46.12) >DroSec_CAF1 110394 120 - 1 CCGCCCCAAAAUCCCAGACCUCCUCCAAUGCCAGGCCAGGUAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUUAGGUGGCCCCUGCCCCCUUU ..(((((((.......((.....))....(((((((..((.....(((((((((((((....))))..))))).))))...))..)))))))))))))).(((.(((....))).))).. ( -43.80) >DroSim_CAF1 122594 120 - 1 CCGCCCCAAAAUCCCAGACCUCCUCCAAUGCCAGGCCAGGUAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUCAGGUGGCCCCUGCCCCCUUU ................((((....((((.(((((((..((.....(((((((((((((....))))..))))).))))...))..)))))))))))))))(((.(((....))).))).. ( -44.40) >DroEre_CAF1 107864 107 - 1 CCCGCUGCAAAUCCCAAACCA---CCAAUGCCAGGCCAGGUAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGU-UGGGGUCAG---------GCCCGCUUU ...((.((..((((((.....---.....(((((((..((.....(((((((((((((....))))..))))).))))...))..)))))))-))))))...---------))..))... ( -35.90) >DroYak_CAF1 110356 117 - 1 CCGCCCCCAAAUCCCAAACCU---CCAAUGCCAGGCCAGGUAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGUUUGGGGUCAUGUGGCCCCUGCCCGCCUU ..((.................---.....(((((((..((.....(((((((((((((....))))..))))).))))...))..)))))))..((((((....)))))).....))... ( -38.10) >consensus CCGCCCCAAAAUCCCAAACCUCCUCCAAUGCCAGGCCAGGUAUAUUUGUCGCCGGUCUUAAGAGAUUGCGGCGCACAAUUUCCGCGCUUGGCUUGGGGUCAGGUGGCCCCUGCCCCCUUU ..(((.(...(((((((............(((((((..((.....(((((((((((((....))))..))))).))))...))..))))))))))))))...).)))............. (-34.54 = -35.34 + 0.80)

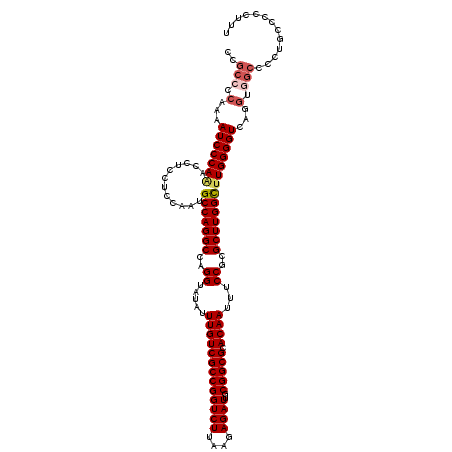
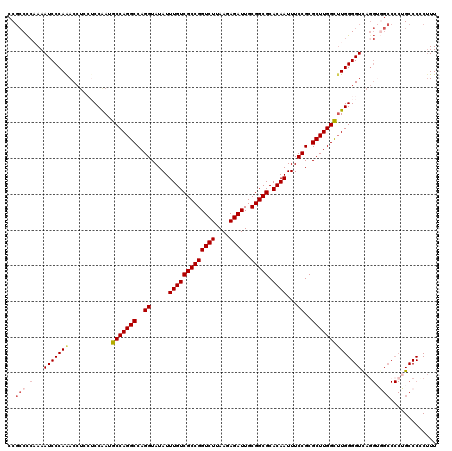
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:45 2006