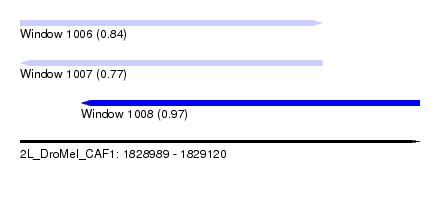
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,828,989 – 1,829,120 |
| Length | 131 |
| Max. P | 0.969076 |

| Location | 1,828,989 – 1,829,088 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -16.89 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1828989 99 + 22407834 ----CGGGGCGGGCCUCU----------------AGUGCCAUUUGUAAAUCAAAUUCUGACUUUGCGAAACAUCAAAAUGCUUCGAGAGCCCCCCUCA-AAAAACGAAAAUUAUUGCCAA ----.((((.(((((((.----------------..........((((((((.....))).)))))(((.(((....))).)))))).)))).)))).-..................... ( -25.00) >DroSec_CAF1 49007 114 + 1 ----CCGGGCGGGCCUCCAAUUCUGGGCCAGCCCAGUGCCAUUUGUAAAUCAAAUUCUGACUCUGCGAAACAUCAAAAUGUUUCGAGAGCCC-CCUCA-AAAAACGAAAAUUAUUGCCAA ----..((((.(((((........))))).)))).......(((((...(((.....)))((((.((((((((....))))))))))))...-.....-....)))))............ ( -32.80) >DroSim_CAF1 29570 114 + 1 ----CGGGGCGAGCCUCCAAUUCUGGGCCAGCCCAGUGCCAUUUGUAAAUCAAAUUCUGACUCUGCGAAACAUCAAAAUGUUUCGAGAGCCC-CCUCA-AAAAAUGAAAAUUAUUGCCAA ----.((((((((.........(((((....)))))....(((((.....))))).....)))..((((((((....))))))))...))))-)....-..................... ( -31.00) >DroEre_CAF1 37564 119 + 1 GCAAUGGGGCGGGCCACCCAUUUUGGGCCAAUGCAGCGCCCUUUGUAAAUCAAAUUCUGGCCCUGCGAAACAUCAAAAUGUUUCCAGAGCGC-CCUGAGAAAAAUGAAAAUUAUUACCAA .....(((((((((((........(((((......).))))((((.....))))...)))))(((.(((((((....))))))))))..)))-)))........................ ( -36.30) >DroYak_CAF1 13979 109 + 1 ----UGGGGCGG-----CAAUUCUGGGCCAACCCAGUGCCAUUUGUAAAUCAAAUUCUGGCCCUGCGAAACAUCAAAAUGUUUCGAGAGCGC-CCUCA-AAAAACGAAAAUUAUUGCCAA ----.(((((((-----((...(((((....)))))))))(((((.....)))))....((.((.((((((((....)))))))))).))))-)))..-..................... ( -35.20) >consensus ____CGGGGCGGGCCUCCAAUUCUGGGCCAACCCAGUGCCAUUUGUAAAUCAAAUUCUGACUCUGCGAAACAUCAAAAUGUUUCGAGAGCCC_CCUCA_AAAAACGAAAAUUAUUGCCAA .......(((((..........(((((....))))).....(((((...(((.....)))((((.((((((((....))))))))))))..............))))).....))))).. (-16.89 = -17.60 + 0.71)

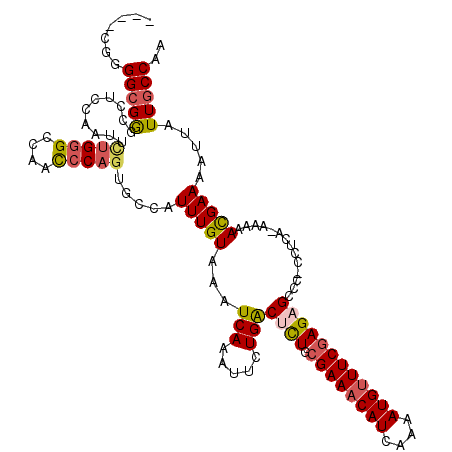
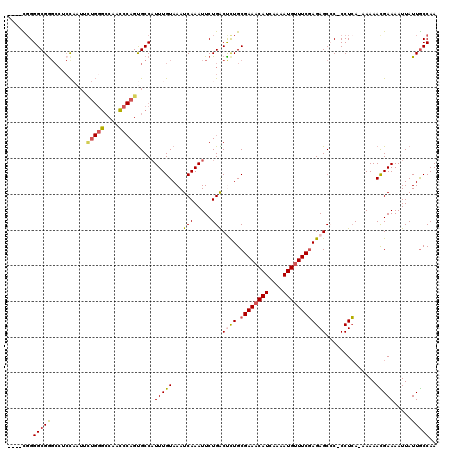
| Location | 1,828,989 – 1,829,088 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -21.35 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1828989 99 - 22407834 UUGGCAAUAAUUUUCGUUUUU-UGAGGGGGGCUCUCGAAGCAUUUUGAUGUUUCGCAAAGUCAGAAUUUGAUUUACAAAUGGCACU----------------AGAGGCCCGCCCCG---- ..(((.....((((((.....-))))))((((.((((((((((....))))))).....((((...((((.....))))))))...----------------.))))))))))...---- ( -29.10) >DroSec_CAF1 49007 114 - 1 UUGGCAAUAAUUUUCGUUUUU-UGAGG-GGGCUCUCGAAACAUUUUGAUGUUUCGCAGAGUCAGAAUUUGAUUUACAAAUGGCACUGGGCUGGCCCAGAAUUGGAGGCCCGCCCGG---- ...((.....((((((.....-)))))-)((((((((((((((....)))))))).))))))...(((((.....))))).)).((((((.(((((.......).)))).))))))---- ( -40.70) >DroSim_CAF1 29570 114 - 1 UUGGCAAUAAUUUUCAUUUUU-UGAGG-GGGCUCUCGAAACAUUUUGAUGUUUCGCAGAGUCAGAAUUUGAUUUACAAAUGGCACUGGGCUGGCCCAGAAUUGGAGGCUCGCCCCG---- ............((((.....-))))(-((((...((((((((....))))))))..(((((.(.(((((.....)))))..).(((((....))))).......)))))))))).---- ( -38.50) >DroEre_CAF1 37564 119 - 1 UUGGUAAUAAUUUUCAUUUUUCUCAGG-GCGCUCUGGAAACAUUUUGAUGUUUCGCAGGGCCAGAAUUUGAUUUACAAAGGGCGCUGCAUUGGCCCAAAAUGGGUGGCCCGCCCCAUUGC ...(((((.................((-((......(((((((....)))))))((((.(((....((((.....)))).))).))))....)))).....(((((...))))).))))) ( -37.80) >DroYak_CAF1 13979 109 - 1 UUGGCAAUAAUUUUCGUUUUU-UGAGG-GCGCUCUCGAAACAUUUUGAUGUUUCGCAGGGCCAGAAUUUGAUUUACAAAUGGCACUGGGUUGGCCCAGAAUUG-----CCGCCCCA---- .....................-((.((-(((((((((((((((....)))))))).)))))....(((((.....)))))(((((((((....)))))...))-----))))))))---- ( -39.20) >consensus UUGGCAAUAAUUUUCGUUUUU_UGAGG_GGGCUCUCGAAACAUUUUGAUGUUUCGCAGAGUCAGAAUUUGAUUUACAAAUGGCACUGGGCUGGCCCAGAAUUGGAGGCCCGCCCCG____ ..(((........(((..(((........((((((((((((((....)))))))).)))))).)))..))).............(((((....)))))............)))....... (-21.35 = -22.26 + 0.91)

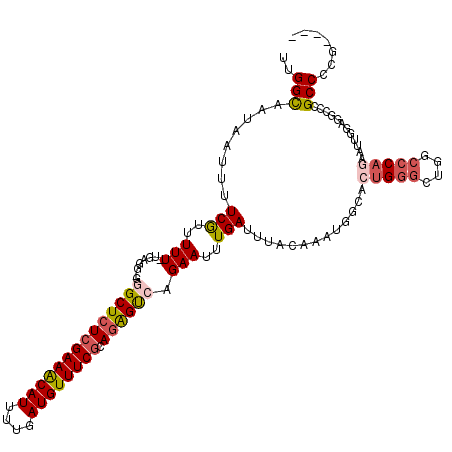
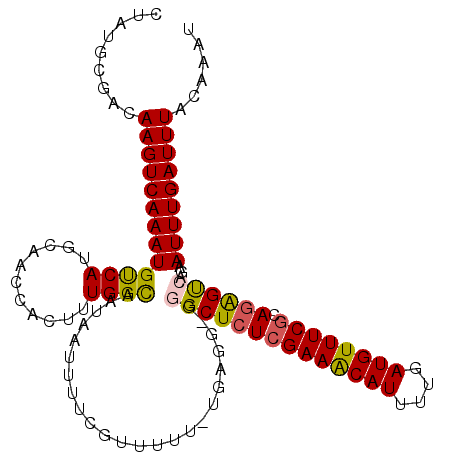
| Location | 1,829,009 – 1,829,120 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1829009 111 - 22407834 CUAUGCGACAAGUCAAAUGUCAUGCAACCACUUUGGCAAUAAUUUUCGUUUUU-UGAGGGGGGCUCUCGAAGCAUUUUGAUGUUUCGCAAAGUCAGAAUUUGAUUUACAAAU ...(((((...((((((....((((..((.....))...............((-((((((...))))))))))))))))))...)))))(((((((...)))))))...... ( -24.80) >DroSec_CAF1 49043 110 - 1 CUAUGCGACAAGUCAAAUGUCAUGCAACCACUUUGGCAAUAAUUUUCGUUUUU-UGAGG-GGGCUCUCGAAACAUUUUGAUGUUUCGCAGAGUCAGAAUUUGAUUUACAAAU .........(((((((((.((......((.(((.(((..........)))...-.))))-)((((((((((((((....)))))))).)))))).)))))))))))...... ( -28.10) >DroSim_CAF1 29606 110 - 1 CUAUACGACAAGUCAAAUGUCAUGCAACCACUUUGGCAAUAAUUUUCAUUUUU-UGAGG-GGGCUCUCGAAACAUUUUGAUGUUUCGCAGAGUCAGAAUUUGAUUUACAAAU .........(((((((((((((...........)))).....((((((.....-)))))-)((((((((((((((....)))))))).))))))...)))))))))...... ( -28.60) >DroEre_CAF1 37604 111 - 1 CUGUGCGACAAGUCAAAUGCCAUGCACCCACUUUGGUAAUAAUUUUCAUUUUUCUCAGG-GCGCUCUGGAAACAUUUUGAUGUUUCGCAGGGCCAGAAUUUGAUUUACAAAG .........((((((((((((.((.........(((.........))).......)).)-))(((((((((((((....))))))).))))))....)))))))))...... ( -26.69) >DroYak_CAF1 14010 110 - 1 CUAUACGACAAGUCAAAUGCCAUGCACCCACUUUGGCAAUAAUUUUCGUUUUU-UGAGG-GCGCUCUCGAAACAUUUUGAUGUUUCGCAGGGCCAGAAUUUGAUUUACAAAU .........((((((((((((.(((..........)))......((((.....-)))))-))(((((((((((((....)))))))).)))))....)))))))))...... ( -29.30) >consensus CUAUGCGACAAGUCAAAUGUCAUGCAACCACUUUGGCAAUAAUUUUCGUUUUU_UGAGG_GGGCUCUCGAAACAUUUUGAUGUUUCGCAGAGUCAGAAUUUGAUUUACAAAU .........(((((((((((((...........))))........................((((((((((((((....)))))))).))))))...)))))))))...... (-24.34 = -24.10 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:03 2006