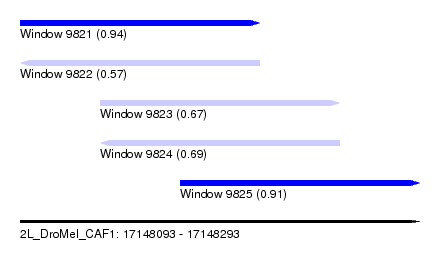
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,148,093 – 17,148,293 |
| Length | 200 |
| Max. P | 0.942734 |

| Location | 17,148,093 – 17,148,213 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -32.02 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17148093 120 + 22407834 UUUGGCCUCAUUAAAGCGCCUCCGGUCAGAGCCGGGCCACUGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCU (((((((....((..(((.((((((((((((.(((....))).((((....)))).)))).))))))....)).)))..)).......(((((((....))))))).)))))))...... ( -37.40) >DroSec_CAF1 108880 120 + 1 UUUGGCCUCAUUAAAGCGCCUCCGGUCAAAGCCGGGCCACUGAAUUAUUUAUAAUACUCUCAACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCU (((((((........((((.((.(((....)))(((((..(((................)))...)))))..)).).)))........(((((((....))))))).)))))))...... ( -32.09) >DroSim_CAF1 121110 120 + 1 UUUGGCCUCAUUAAAGCGCCUCCGGUCAGAGCCGGGCCACUGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACUUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCU (((((((......((((....((((((((((.(((....))).((((....)))).)))).)))))).....).)))...........(((((((....))))))).)))))))...... ( -35.80) >DroEre_CAF1 106479 120 + 1 UUUGGCCUCAUUAAAGCGCCUCCGGUCAGAGCCGGGCCACUGAAUUAUUUAUAAUACUCUCGGCCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUGUGCCACGGCCAAAUUGUCU (((((((....((..(((.((((((((((((.(((....))).((((....)))).)))).))))))....)).)))..)).......((((((......)))))).)))))))...... ( -36.10) >DroYak_CAF1 108984 120 + 1 UUUGGCCUCAUUAAAGUGCCUCCGGUCUGAGCCGGGCCACUGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUGUGCCACGGCCAAAUUGUCU (((((((.......((((.(.(((((....)))))).))))...................((((..(((...))))))).........((((((......)))))).)))))))...... ( -34.20) >consensus UUUGGCCUCAUUAAAGCGCCUCCGGUCAGAGCCGGGCCACUGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCU (((((((....((..(((.((((((((((((.(((....))).((((....)))).)))).))))))....)).)))..)).......(((((((....))))))).)))))))...... (-32.02 = -32.30 + 0.28)

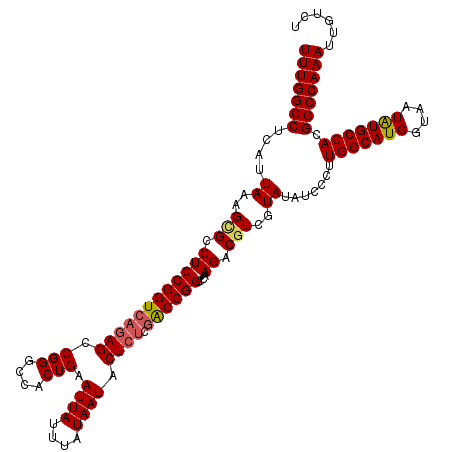
| Location | 17,148,093 – 17,148,213 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -34.56 |
| Energy contribution | -34.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17148093 120 - 22407834 AGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCAGUGGCCCGGCUCUGACCGGAGGCGCUUUAAUGAGGCCAAA ......(((((((.((((((......))))))..........(.(((((....((((((.(((((.......((......))......)))))))))))))))))........))))))) ( -38.92) >DroSec_CAF1 108880 120 - 1 AGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUUGAGAGUAUUAUAAAUAAUUCAGUGGCCCGGCUUUGACCGGAGGCGCUUUAAUGAGGCCAAA ......(((((((.((((((......)))))).....((((..((.(((..(.....)..)))))..))))........((((.(((((......)))).).)))).......))))))) ( -34.70) >DroSim_CAF1 121110 120 - 1 AGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGAAGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCAGUGGCCCGGCUCUGACCGGAGGCGCUUUAAUGAGGCCAAA ......(((((((.((((((......))))))..........((((((((...((((((.(((((.......((......))......)))))))))))))).))))).....))))))) ( -37.42) >DroEre_CAF1 106479 120 - 1 AGACAAUUUGGCCGUGGCACAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGCCGAGAGUAUUAUAAAUAAUUCAGUGGCCCGGCUCUGACCGGAGGCGCUUUAAUGAGGCCAAA ......(((((((.(((((........))))).....((((..((.((((((.....))))))))..))))........((((.(((((......)))).).)))).......))))))) ( -37.30) >DroYak_CAF1 108984 120 - 1 AGACAAUUUGGCCGUGGCACAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCAGUGGCCCGGCUCAGACCGGAGGCACUUUAAUGAGGCCAAA ......(((((((((((.......))))(((..(((((.......)))))...((((((..((((.......((......))......)))).)))))).)))..........))))))) ( -35.72) >consensus AGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCAGUGGCCCGGCUCUGACCGGAGGCGCUUUAAUGAGGCCAAA ......(((((((.((((((......)))))).....((((..((.((((((.....))))))))..))))........((((.(((((......)))).).)))).......))))))) (-34.56 = -34.68 + 0.12)

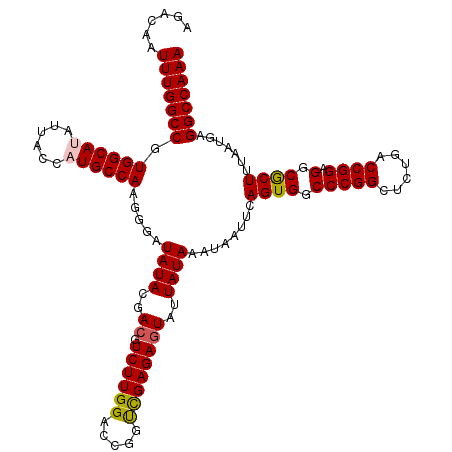
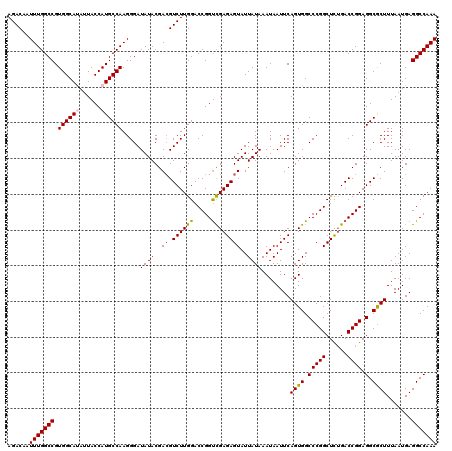
| Location | 17,148,133 – 17,148,253 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -24.18 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17148133 120 + 22407834 UGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAGUAUAUAUAUACACCUCCCUCUGAGGUGAGUGUCC .............((((((..(((..(((...))))))((((((.((.(((((((....))))))).))....(((((....)))))....))))))(((((.....))))))))))).. ( -28.30) >DroSec_CAF1 108920 119 + 1 UGAAUUAUUUAUAAUACUCUCAACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCUCGCUCUGAGGUGAGUGUCC .....(((((((((.((......((((((...))).............(((((((....)))))))))).......))..))))))-))).((((.((((((.....)))))).)))).. ( -28.02) >DroSim_CAF1 121150 119 + 1 UGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACUUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCUCGCUCUGAGGUGAGUGUCC .....(((((((((.((...(((..((((...)))))))......((.(((((((....))))))).)).......))..))))))-))).((((.((((((.....)))))).)))).. ( -29.10) >DroEre_CAF1 106519 119 + 1 UGAAUUAUUUAUAAUACUCUCGGCCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUGUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCGCCCUCUGAGGUGAGCGUCC .....(((((((((.((....((((((((...))).............((((((......))))))))))).....))..))))))-)))......((((.........))))....... ( -29.30) >DroYak_CAF1 109024 119 + 1 UGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUGUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCUCCCUCGGAGGUGAGUGUCC .....(((((((((.((...((((..(((...)))))))......((.((((((......)))))).)).......))..))))))-))).((((.(((((((...))))))).)))).. ( -31.60) >consensus UGAAUUAUUUAUAAUACUCUCGACCGGUCCAAGACGUCGUAUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAA_AUAUAUAUACACCUCCCUCUGAGGUGAGUGUCC .............((((((...........((((((.........((.(((((((....))))))).))......))))))................(((((.....))))))))))).. (-24.18 = -24.54 + 0.36)

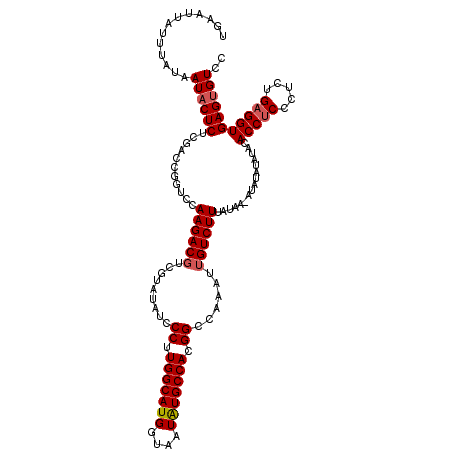
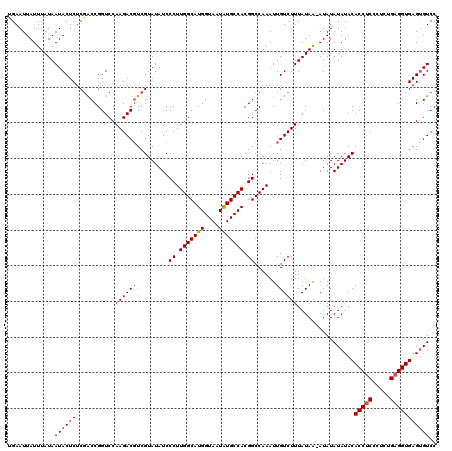
| Location | 17,148,133 – 17,148,253 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -27.56 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17148133 120 - 22407834 GGACACUCACCUCAGAGGGAGGUGUAUAUAUAUACUAUAAAGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCA ..(((((..((.....))..))))).((((.(((((.....(((....(((((.((((((......)))))).(((((.......))))))).))))))...)))))))))......... ( -29.10) >DroSec_CAF1 108920 119 - 1 GGACACUCACCUCAGAGCGAGGUGUAUAUAUAU-UUAUAAAGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUUGAGAGUAUUAUAAAUAAUUCA (((....((((((.....))))))......(((-((((((..((..((..((((((((((......)))))).(((((.......)))))....))))..)).)).))))))))).))). ( -30.90) >DroSim_CAF1 121150 119 - 1 GGACACUCACCUCAGAGCGAGGUGUAUAUAUAU-UUAUAAAGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGAAGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCA (((....((((((.....))))))......(((-((((((..((..((((((((((((((......)))))).(((((.......)))))....)))))))).)).))))))))).))). ( -32.40) >DroEre_CAF1 106519 119 - 1 GGACGCUCACCUCAGAGGGCGGUGUAUAUAUAU-UUAUAAAGACAAUUUGGCCGUGGCACAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGCCGAGAGUAUUAUAAAUAAUUCA ((((((((........))))).........(((-((((((..((..(((((((((((((........))))).(((((.......)))))....)))))))).)).))))))))).))). ( -31.80) >DroYak_CAF1 109024 119 - 1 GGACACUCACCUCCGAGGGAGGUGUAUAUAUAU-UUAUAAAGACAAUUUGGCCGUGGCACAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCA (((....(((((((...)))))))......(((-((((((..((..(((((((((((((........))))).(((((.......)))))....)))))))).)).))))))))).))). ( -32.40) >consensus GGACACUCACCUCAGAGGGAGGUGUAUAUAUAU_UUAUAAAGACAAUUUGGCCGUGGCAUAUUACCAUGCCAAGGGAUAUACGACGUCUUGGACCGGUCGAGAGUAUUAUAAAUAAUUCA ....(((((((((.....)))))........................(((((((((((((......)))))).(((((.......)))))....)))))))))))............... (-27.56 = -27.68 + 0.12)

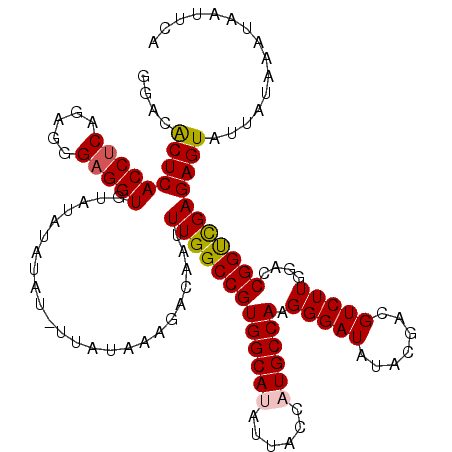
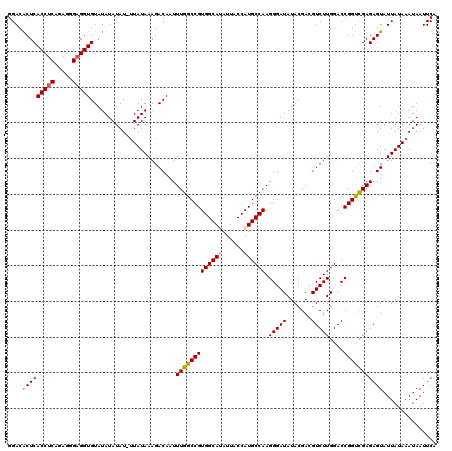
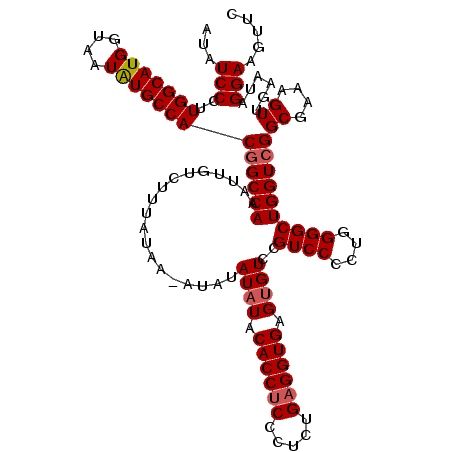
| Location | 17,148,173 – 17,148,293 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -35.82 |
| Energy contribution | -36.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17148173 120 + 22407834 AUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAGUAUAUAUAUACACCUCCCUCUGAGGUGAGUGUCCGUCCCCUGGGGCUGGUCGGCGAAAGUUGAAUAGGAAGUUC ...(((..(((((((....)))))))((((((...................((((.((((((.....)))))).))))..((((....))))))))))((....))......)))..... ( -38.20) >DroSec_CAF1 108960 119 + 1 AUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCUCGCUCUGAGGUGAGUGUCCGUCCCCUGGGGCUGGUAGGCGAAAGUUGAAUAGGAAGUUC ...(((.(((((.((((.....))))..))))).((((((......-....((((.((((((.....)))))).))))((((((....)))).)).))))))..........)))..... ( -35.10) >DroSim_CAF1 121190 119 + 1 AUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCUCGCUCUGAGGUGAGUGUCCGUCCCCUGGGGCUGGUCGGCGAAAGUUGAAUAGGAAGUUC ...(((..(((((((....)))))))((((((..............-....((((.((((((.....)))))).))))..((((....))))))))))((....))......)))..... ( -38.50) >DroEre_CAF1 106559 119 + 1 AUAUCCCUUGGCAUGGUAAUGUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCGCCCUCUGAGGUGAGCGUCCGUCCCCUGGGGCUGGUCGGCGAAAGUUGAAUAGGAAGUUC ...(((.(((((.((((.....))))..))))).............-...........(((((((...(((.(..(....)..))))))))).))(((((....)))))...)))..... ( -33.90) >DroYak_CAF1 109064 119 + 1 AUAUCCCUUGGCAUGGUAAUGUGCCACGGCCAAAUUGUCUUUAUAA-AUAUAUAUACACCUCCCUCGGAGGUGAGUGUCCGUCCCCUGGGGCUGGUCGGCGAAAGUUGAAUAGGAAGUUC ...(((..((((((......))))))((((((..............-....((((.(((((((...))))))).))))..((((....))))))))))((....))......)))..... ( -39.90) >consensus AUAUCCCUUGGCAUGGUAAUAUGCCACGGCCAAAUUGUCUUUAUAA_AUAUAUAUACACCUCCCUCUGAGGUGAGUGUCCGUCCCCUGGGGCUGGUCGGCGAAAGUUGAAUAGGAAGUUC ...(((..(((((((....)))))))((((((...................((((.((((((.....)))))).))))..((((....))))))))))((....))......)))..... (-35.82 = -36.18 + 0.36)

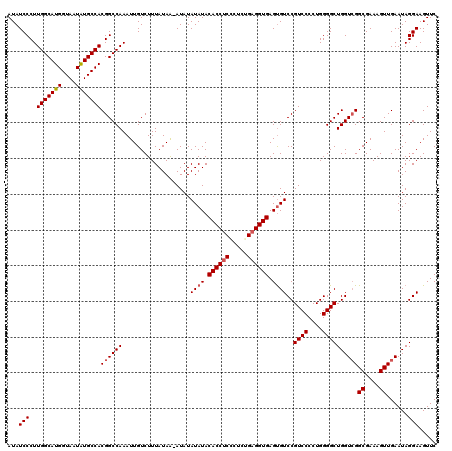
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:41 2006