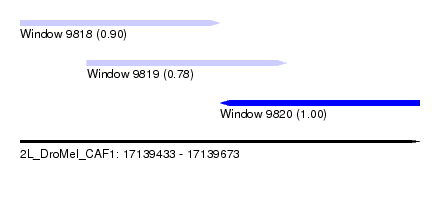
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,139,433 – 17,139,673 |
| Length | 240 |
| Max. P | 0.999509 |

| Location | 17,139,433 – 17,139,553 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.18 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17139433 120 + 22407834 UUCCUGGCCUUCAUUUUUCAACUUCAACGACAGUGGCAGCCGCGUGUCUGUGGCUCCUGCCUCAUGGCUUGCGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUU ...((((.....................((((((((...)))).))))((((((....(((....)))(((((.....))))).......)))))).......))))............. ( -30.20) >DroSec_CAF1 107845 120 + 1 UUCCUGGCCUUCAUUUUUCAACUUCAACGACAGUGGCAGCCGCGUGUCUGUGGCUCCUGCCGCAUGGCUUGUGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUU ...((((..........((.........))..(((((((.((((.(((((((((....)))))).))).))))((((......)))).)))))))........))))............. ( -31.10) >DroSim_CAF1 120073 120 + 1 UUCCUGGCCUUCAUUUUUCAACUUCAACGACAGUGGCAGCCGCGUGUCUGUGGCUCCUGCCGCAUGGCUUGUGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUU ...((((..........((.........))..(((((((.((((.(((((((((....)))))).))).))))((((......)))).)))))))........))))............. ( -31.10) >DroEre_CAF1 105452 117 + 1 UUCCUGGCCUUCAUUUUUCAACUUCAACGACAGUGGGAGCCGCGUGUCUGUGGCUCCUGCCUCAUGGCU---GGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCUAGCACUUUUUGUUUU ...((((.....................((..(..(((((((((....)))))))))..).)).((((.---(((((......)))))..)))).........))))............. ( -31.00) >DroYak_CAF1 107927 120 + 1 UUCCUGGCCUUCAUUUUUCAACUUCAACGACAGUGGGAGCCGCGUGUCUGUGGCUUCUGCCUCAUGCCUUGCGGAAUAUGCAAAUUCUUUGCCACAAUUUUUUCUGGCACUUUUUGUUUU ((((.(((..(((((..((.........)).)))))..)))(((.((.((.(((....))).)).))..)))))))...(((((.....(((((.((....)).)))))...)))))... ( -29.70) >consensus UUCCUGGCCUUCAUUUUUCAACUUCAACGACAGUGGCAGCCGCGUGUCUGUGGCUCCUGCCUCAUGGCUUGCGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUU ...((((.....................((((((((...)))).))))((((((....(((....)))....(((((......)))))..)))))).......))))............. (-25.38 = -25.18 + -0.20)

| Location | 17,139,473 – 17,139,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17139473 120 + 22407834 CGCGUGUCUGUGGCUCCUGCCUCAUGGCUUGCGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUUUAUCCUGUUGCUGUUGUCGAAGUCGUCUUUGAUAAGCAAA .((.....((((((....(((....)))(((((.....))))).......))))))........((((((................).)))))(((((((((....)))))))))))... ( -31.99) >DroSec_CAF1 107885 120 + 1 CGCGUGUCUGUGGCUCCUGCCGCAUGGCUUGUGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUUUAUCCUGUUGCUGUUGUCGAAGUCGUCUUUGAUAAGCAAA ...((((.((((((....))))))((((....(((((......)))))..))))............)))).................(((((..((((((((....))))))))))))). ( -32.90) >DroSim_CAF1 120113 120 + 1 CGCGUGUCUGUGGCUCCUGCCGCAUGGCUUGUGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUUUAUCCUGUUGCUGUUGUCGAAGUCGUCUUUGAUAAGCAAA ...((((.((((((....))))))((((....(((((......)))))..))))............)))).................(((((..((((((((....))))))))))))). ( -32.90) >DroEre_CAF1 105492 117 + 1 CGCGUGUCUGUGGCUCCUGCCUCAUGGCU---GGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCUAGCACUUUUUGUUUUUAUCCUGUUGCUGUUGCCGAAGUCGUCUUUGAUAAGCAAA ...((((.((.(((....))).))((((.---(((((......)))))..))))............)))).................(((((.....(((((....)))))...))))). ( -25.70) >DroYak_CAF1 107967 120 + 1 CGCGUGUCUGUGGCUUCUGCCUCAUGCCUUGCGGAAUAUGCAAAUUCUUUGCCACAAUUUUUUCUGGCACUUUUUGUUUUCAUCCUGUUGCUGUUGCCGAAGUCGUCUUUGAUAAGCAAA .((.((((.((((((((.((..((.((...(((((....(((((.....(((((.((....)).)))))...))))).....))).)).))))..)).))))))).)...)))).))... ( -32.40) >consensus CGCGUGUCUGUGGCUCCUGCCUCAUGGCUUGCGGAAUAUGCAAAUUCUUUGCCACAAUUUUCCCCAGCACUUUUUGUUUUUAUCCUGUUGCUGUUGUCGAAGUCGUCUUUGAUAAGCAAA ...(((.(((.(((....)))...((((....(((((......)))))..))))..........)))))).................(((((..((((((((....))))))))))))). (-26.08 = -26.28 + 0.20)


| Location | 17,139,553 – 17,139,673 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17139553 120 - 22407834 AUCAAAUAGCUCAGGCCUUUUCUUCUAAUGCUUUUCGAGCUCGUCAACGCACUAUAAAAAUAUCAUUAUCAGUGCAAAAGUUUGCUUAUCAAAGACGACUUCGACAACAGCAACAGGAUA ........((....))......((((..((((..(((((.(((((...(((((((((........)))).)))))..(((....)))......))))).)))))....))))..)))).. ( -26.30) >DroSec_CAF1 107965 120 - 1 AUCAAAUAACUCAGGCCUUUUCUCCUAAUGCUUUGCGAGCUCGUCAACGCACUAUAAAAAUAUCAUUAUCAGUGCAAAAGUUUGCUUAUCAAAGACGACUUCGACAACAGCAACAGGAUA ......................((((..(((((((((((.(((((...(((((((((........)))).)))))..(((....)))......))))).)))).))).))))..)))).. ( -30.30) >DroSim_CAF1 120193 120 - 1 AUCAAAUAACUCAGGCCUUUUCUCCUAAUGCUUUGCGAGCUCGUCAACGCACUAUAAAAAUAUCAUUAUCAGAGCAAAAGUUUGCUUAUCAAAGACGACUUCGACAACAGCAACAGGAUA ......................((((..(((((((((((.(((((..............(((....)))..((((((....))))))......))))).)))).))).))))..)))).. ( -25.70) >DroEre_CAF1 105569 120 - 1 GUCAAAUAUCGCAGGCCUCUUCUUCUAAUGCUUUUCGAGCUCGUCAACGCACUAUAAAAAUAUCAUUAUCAGUGCAAAAGUUUGCUUAUCAAAGACGACUUCGGCAACAGCAACAGGAUA ......................((((..((((..(((((.(((((...(((((((((........)))).)))))..(((....)))......))))).)))))....))))..)))).. ( -24.80) >DroYak_CAF1 108047 120 - 1 ACCAAAUAUCACAGGUCUCUUCUCAUCUUGCUUUUCGAGCUCGUCAACGCACUAUAAAAAUAUCAUUAUCAGUGCAAAAGUUUGCUUAUCAAAGACGACUUCGGCAACAGCAACAGGAUG (((..........))).......(((((((.....((((.(((((...(((((((((........)))).)))))..(((....)))......))))).))))((....))..))))))) ( -27.10) >consensus AUCAAAUAACUCAGGCCUUUUCUCCUAAUGCUUUUCGAGCUCGUCAACGCACUAUAAAAAUAUCAUUAUCAGUGCAAAAGUUUGCUUAUCAAAGACGACUUCGACAACAGCAACAGGAUA ......................((((..((((...((((.(((((...(((((((((........)))).)))))..(((....)))......))))).)))).....))))..)))).. (-23.94 = -24.10 + 0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:36 2006