
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,136,971 – 17,137,207 |
| Length | 236 |
| Max. P | 0.875888 |

| Location | 17,136,971 – 17,137,090 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136971 119 + 22407834 UAACUUUGAUGUCGCCGAAAGUCUCUUUGCUCUCUUUUAUUUAUUACACGCCCGCAGAUGCCCCGUUUCGUUAUAUUCGGUGGAAAAUGGGCGAACUACUUUUCGGCUUUUGGCGUUGU ............(((((((((((..........................(((((......(((((............))).))....)))))(((......)))))))))))))).... ( -31.00) >DroSec_CAF1 105335 118 + 1 UAACUUUGAUGUCGCCGAAAGUCUCUUUGCUCUCUUUUAUGUAAUA-ACGCCCUCAGAUGCCCCGUUUCGUUAUAUUCGGUGGAAAAUGGGCGAACUACUUUUCGGCUUUUGGCGUUGU ............(((((((((((...((((..........))))..-.(((((.......(((((............))).)).....)))))...........))))))))))).... ( -32.90) >DroSim_CAF1 117551 119 + 1 UAACUUUGAUGUCGCCGAAAGUCUCUUUGCUCUCUUUUAUUUAUUACACGCCCGCAGAUGCCCCGUUUCGUUAUAUUCGGUGGAAAAUGGGCGAACUACUUUUCGGCUUUUGGCGUUGU ............(((((((((((..........................(((((......(((((............))).))....)))))(((......)))))))))))))).... ( -31.00) >DroEre_CAF1 102897 119 + 1 UAACUUUGAUGUCGCCGAAAGUCUCUGUGCUCUCUUUUAUUUAUUACACGCCCGCAGAUGCCCCGUUUCGUUAUAUUCGGUGGAAAAUGGGCGAACUACUUUUCGGCUUUUGGCGUUGU ............(((((((((((..((((...............))))((((((......(((((............))).))....))))))...........))))))))))).... ( -31.56) >DroYak_CAF1 105396 119 + 1 UAACUUUGAUGUCGCCGAAAGUCUCUUUGCUCUCUUUUAUUUAUUACACGCCCGCAGAUGCCCCGUUUCGUUAUAUUCGGUGGAAAAUGGGCGAACUACUUUUCGGCUUUUGGCGUUGU ............(((((((((((..........................(((((......(((((............))).))....)))))(((......)))))))))))))).... ( -31.00) >consensus UAACUUUGAUGUCGCCGAAAGUCUCUUUGCUCUCUUUUAUUUAUUACACGCCCGCAGAUGCCCCGUUUCGUUAUAUUCGGUGGAAAAUGGGCGAACUACUUUUCGGCUUUUGGCGUUGU ............(((((((((((..........................(((((......(((((............))).))....)))))(((......)))))))))))))).... (-30.48 = -30.68 + 0.20)

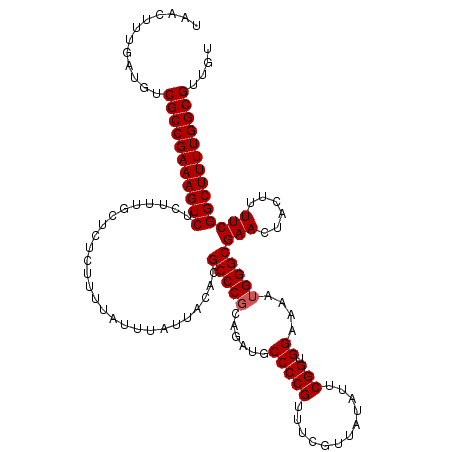

| Location | 17,136,971 – 17,137,090 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136971 119 - 22407834 ACAACGCCAAAAGCCGAAAAGUAGUUCGCCCAUUUUCCACCGAAUAUAACGAAACGGGGCAUCUGCGGGCGUGUAAUAAAUAAAAGAGAGCAAAGAGACUUUCGGCGACAUCAAAGUUA ....((((....(((.....((((...((((..((((.............))))..))))..)))).)))...............(((((........)))))))))............ ( -28.22) >DroSec_CAF1 105335 118 - 1 ACAACGCCAAAAGCCGAAAAGUAGUUCGCCCAUUUUCCACCGAAUAUAACGAAACGGGGCAUCUGAGGGCGU-UAUUACAUAAAAGAGAGCAAAGAGACUUUCGGCGACAUCAAAGUUA ....((((............(((((.(((((.....((.(((............))))).......))))).-.)))))......(((((........)))))))))............ ( -28.90) >DroSim_CAF1 117551 119 - 1 ACAACGCCAAAAGCCGAAAAGUAGUUCGCCCAUUUUCCACCGAAUAUAACGAAACGGGGCAUCUGCGGGCGUGUAAUAAAUAAAAGAGAGCAAAGAGACUUUCGGCGACAUCAAAGUUA ....((((....(((.....((((...((((..((((.............))))..))))..)))).)))...............(((((........)))))))))............ ( -28.22) >DroEre_CAF1 102897 119 - 1 ACAACGCCAAAAGCCGAAAAGUAGUUCGCCCAUUUUCCACCGAAUAUAACGAAACGGGGCAUCUGCGGGCGUGUAAUAAAUAAAAGAGAGCACAGAGACUUUCGGCGACAUCAAAGUUA ....((((....(((.....((((...((((..((((.............))))..))))..)))).)))...............(((((........)))))))))............ ( -28.22) >DroYak_CAF1 105396 119 - 1 ACAACGCCAAAAGCCGAAAAGUAGUUCGCCCAUUUUCCACCGAAUAUAACGAAACGGGGCAUCUGCGGGCGUGUAAUAAAUAAAAGAGAGCAAAGAGACUUUCGGCGACAUCAAAGUUA ....((((....(((.....((((...((((..((((.............))))..))))..)))).)))...............(((((........)))))))))............ ( -28.22) >consensus ACAACGCCAAAAGCCGAAAAGUAGUUCGCCCAUUUUCCACCGAAUAUAACGAAACGGGGCAUCUGCGGGCGUGUAAUAAAUAAAAGAGAGCAAAGAGACUUUCGGCGACAUCAAAGUUA ....((((....(((.....((((...((((..((((.............))))..))))..)))).)))...............(((((........)))))))))............ (-27.34 = -27.54 + 0.20)

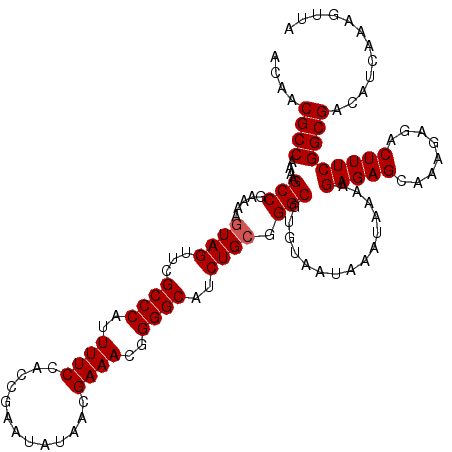

| Location | 17,137,090 – 17,137,207 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 99.32 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17137090 117 + 22407834 UAACAUUUCAUUUCUGCCGCAUUUCAUUUCGCCUGCGAUGCGUUUUAUGAAUUCAGUUAUUCCGUUGGCUGCAUAUUCAAUUU-UUUAUUCGCUUUCCCACCGCAGCUUGGCAUCUCC .......(((...((((((((((.((.......)).)))))).....(((((.((((((......))))))...)))))....-..................))))..)))....... ( -20.50) >DroSec_CAF1 105453 117 + 1 UAACAUUUCAUUUCUGCCGCAUUUCAUUUCGCCUGCGAUGCGUUUUAUGAAUUCAGUUAUUCCGUUGGCUGCAUAUUCAAUUU-UUUAUUCGCUUUCCCACCGCAGCUUGGCAUCUCC .......(((...((((((((((.((.......)).)))))).....(((((.((((((......))))))...)))))....-..................))))..)))....... ( -20.50) >DroSim_CAF1 117670 117 + 1 UAACAUUUCAUUUCUGCCGCAUUUCAUUUCGCCUGCGAUGCGUUUUAUGAAUUCAGUUAUUCCGUUGGCUGCAUAUUCAAUUU-UUUAUUCGCUUUCCCACCGCAGCUUGGCAUCUCC .......(((...((((((((((.((.......)).)))))).....(((((.((((((......))))))...)))))....-..................))))..)))....... ( -20.50) >DroEre_CAF1 103016 117 + 1 UAACAUUUCAUUUCUGCCGCAUUUCAUUUCGCCUGCGAUGCGUUUUAUGAAUUCAGUUAUUCCGUUGGCUGCAUAUUCAAUUU-UUUAUUCGCUUUCCCACCGCAGCUUGGCAUCUCC .......(((...((((((((((.((.......)).)))))).....(((((.((((((......))))))...)))))....-..................))))..)))....... ( -20.50) >DroYak_CAF1 105515 118 + 1 UAACAUUUCAUUUCUGCCGCAUUUCAUUUCGCCUGCGAUGCGUUUUAUGAAUUCAGUUAUUCCGUUGGCUGCAUAUUCAAUUUUUUUAUUCGCUUUCCCACCGCAGCUUGGCAUUUCC .......(((...((((((((((.((.......)).)))))).....(((((.((((((......))))))...))))).......................))))..)))....... ( -20.50) >consensus UAACAUUUCAUUUCUGCCGCAUUUCAUUUCGCCUGCGAUGCGUUUUAUGAAUUCAGUUAUUCCGUUGGCUGCAUAUUCAAUUU_UUUAUUCGCUUUCCCACCGCAGCUUGGCAUCUCC .......(((...((((((((((.((.......)).)))))).....(((((.((((((......))))))...))))).......................))))..)))....... (-20.50 = -20.50 + 0.00)

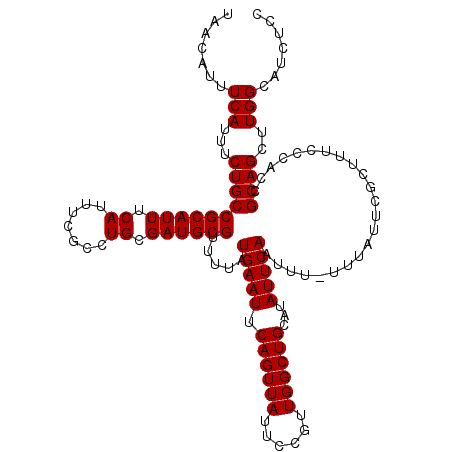
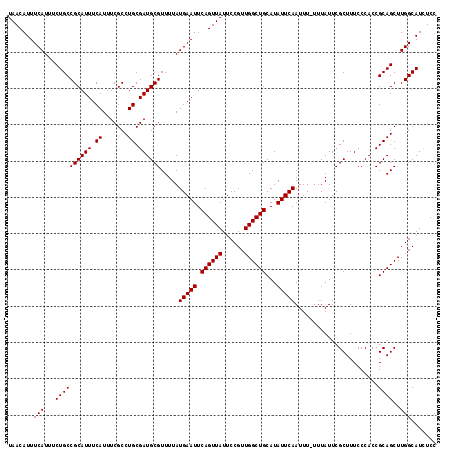
| Location | 17,137,090 – 17,137,207 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 99.32 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17137090 117 - 22407834 GGAGAUGCCAAGCUGCGGUGGGAAAGCGAAUAAA-AAAUUGAAUAUGCAGCCAACGGAAUAACUGAAUUCAUAAAACGCAUCGCAGGCGAAAUGAAAUGCGGCAGAAAUGAAAUGUUA .....((((...((((((((((...(((.((...-.......)).)))..))....((((......))))........))))))))(((........))))))).............. ( -24.10) >DroSec_CAF1 105453 117 - 1 GGAGAUGCCAAGCUGCGGUGGGAAAGCGAAUAAA-AAAUUGAAUAUGCAGCCAACGGAAUAACUGAAUUCAUAAAACGCAUCGCAGGCGAAAUGAAAUGCGGCAGAAAUGAAAUGUUA .....((((...((((((((((...(((.((...-.......)).)))..))....((((......))))........))))))))(((........))))))).............. ( -24.10) >DroSim_CAF1 117670 117 - 1 GGAGAUGCCAAGCUGCGGUGGGAAAGCGAAUAAA-AAAUUGAAUAUGCAGCCAACGGAAUAACUGAAUUCAUAAAACGCAUCGCAGGCGAAAUGAAAUGCGGCAGAAAUGAAAUGUUA .....((((...((((((((((...(((.((...-.......)).)))..))....((((......))))........))))))))(((........))))))).............. ( -24.10) >DroEre_CAF1 103016 117 - 1 GGAGAUGCCAAGCUGCGGUGGGAAAGCGAAUAAA-AAAUUGAAUAUGCAGCCAACGGAAUAACUGAAUUCAUAAAACGCAUCGCAGGCGAAAUGAAAUGCGGCAGAAAUGAAAUGUUA .....((((...((((((((((...(((.((...-.......)).)))..))....((((......))))........))))))))(((........))))))).............. ( -24.10) >DroYak_CAF1 105515 118 - 1 GGAAAUGCCAAGCUGCGGUGGGAAAGCGAAUAAAAAAAUUGAAUAUGCAGCCAACGGAAUAACUGAAUUCAUAAAACGCAUCGCAGGCGAAAUGAAAUGCGGCAGAAAUGAAAUGUUA .....((((...((((((((((...(((.((...........)).)))..))....((((......))))........))))))))(((........))))))).............. ( -24.00) >consensus GGAGAUGCCAAGCUGCGGUGGGAAAGCGAAUAAA_AAAUUGAAUAUGCAGCCAACGGAAUAACUGAAUUCAUAAAACGCAUCGCAGGCGAAAUGAAAUGCGGCAGAAAUGAAAUGUUA .....((((...((((((((((...(((.((...........)).)))..))....((((......))))........))))))))(((........))))))).............. (-24.00 = -24.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:29 2006