
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,136,066 – 17,136,261 |
| Length | 195 |
| Max. P | 0.993829 |

| Location | 17,136,066 – 17,136,181 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.01 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136066 115 + 22407834 CGUCACAUCCAUAACAAUAAAGUCGCAUUAGCAGCUGGGCCAGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUAUUAAUGGUUAUUUUCUUU-----UUAUU ...............(((((((..((....))(((((...)))))..........(((((((.....)))))))......((((((((.......)))))))).....))-----))))) ( -22.20) >DroSec_CAF1 104454 115 + 1 CGUCACAUCCAUAACAAUAAAGUCACAUUAGCAGCUGGUCCCGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU-----UUAUU ..............................(.(((.(....)))).)(((((...(((((((.....)))))))......((((((((.......))))))))....)))-----))... ( -21.00) >DroSim_CAF1 116647 115 + 1 CGUCACAUCCAUAACAAUAAAGUCGCAUUAGCAGCUGGGCCAGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU-----UUAUU ........................((....))(((((...)))))..(((((...(((((((.....)))))))......((((((((.......))))))))....)))-----))... ( -24.60) >DroEre_CAF1 102036 114 + 1 CGUCACAUCCAUAACAAUAAAGUCGCAUUAGCAGCUGGCCCAGCAACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU-----UUAU- ........................((....)).((((...))))...(((((...(((((((.....)))))))......((((((((.......))))))))....)))-----))..- ( -23.90) >DroYak_CAF1 104478 119 + 1 CGUCACAUCCAUAACAAUAAAGUCGCAUUAGCAGCUGGCCCAGCUACAAAACAAUGCUAGCCAAAACAGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUGUUUUCGUUUCGGUUUAU- ........((..(((............((((((((((.....((((((......)).)))).....))))).)))))...((((((((.......))))))))....)))..)).....- ( -23.80) >consensus CGUCACAUCCAUAACAAUAAAGUCGCAUUAGCAGCUGGCCCAGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU_____UUAUU ..............................(.(((((...))))).)........(((((((.....)))))))......((((((((.......))))))))................. (-18.22 = -18.66 + 0.44)



| Location | 17,136,066 – 17,136,181 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.01 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136066 115 - 22407834 AAUAA-----AAAGAAAAUAACCAUUAAUAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCUGGCCCAGCUGCUAAUGCGACUUUAUUGUUAUGGAUGUGACG .....-----((((...(((((((((.....)))))))))......((((.((.....)).)))).((((...(((((((...)))))))....))))))))...(((((....))))). ( -30.20) >DroSec_CAF1 104454 115 - 1 AAUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCGGGACCAGCUGCUAAUGUGACUUUAUUGUUAUGGAUGUGACG .((((-----(((((..(((((((((.....)))))))))......((((.((.....)).)))).)))))))))(((((......)))))...((.(((((((....))))).)).)). ( -31.30) >DroSim_CAF1 116647 115 - 1 AAUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCUGGCCCAGCUGCUAAUGCGACUUUAUUGUUAUGGAUGUGACG ...((-----(((((..(((((((((.....)))))))))......((((.((.....)).)))).)))))))(((((((...)))))))...............(((((....))))). ( -30.90) >DroEre_CAF1 102036 114 - 1 -AUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUUGCUGGGCCAGCUGCUAAUGCGACUUUAUUGUUAUGGAUGUGACG -..((-----(((((..(((((((((.....)))))))))......((((.((.....)).)))).)))))))(((((..(((.....)))...)))))......(((((....))))). ( -31.20) >DroYak_CAF1 104478 119 - 1 -AUAAACCGAAACGAAAACAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACUGUUUUGGCUAGCAUUGUUUUGUAGCUGGGCCAGCUGCUAAUGCGACUUUAUUGUUAUGGAUGUGACG -.....(((.(((((....(((((((.....)))))))......(((((((.....)))))))((((((....(((((((...))))))))))))).......))))).)))........ ( -32.00) >consensus AAUAA_____AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCUGGCCCAGCUGCUAAUGCGACUUUAUUGUUAUGGAUGUGACG .................(((((((((.....)))))))))....(((((.........)))))((((((....(((((((...))))))))))))).........(((((....))))). (-25.70 = -26.14 + 0.44)



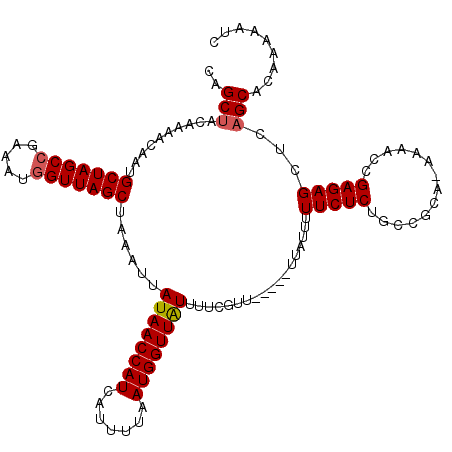
| Location | 17,136,106 – 17,136,221 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136106 115 + 22407834 CAGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUAUUAAUGGUUAUUUUCUUU-----UUAUUUUUCUCUGCCGCACAAAACCGAGAGCUCAGCACAAAAAUC ..(((..........(((((((.....)))))))......((((((((.......)))))))).......-----......(((((..............)))))...)))......... ( -21.14) >DroSec_CAF1 104494 114 + 1 CCGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU-----UUAUUUUUCUCUGCCGCA-AAAACCGAGAGCUCAGCACAAAAAUC ..(((..(((((...(((((((.....)))))))......((((((((.......))))))))....)))-----))....(((((.......-......)))))...)))......... ( -24.52) >DroSim_CAF1 116687 114 + 1 CAGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU-----UUAUUUUUCUCUGCCGCA-AAAACCGAGAGCUCAGCACAAAAAUC ..(((..(((((...(((((((.....)))))))......((((((((.......))))))))....)))-----))....(((((.......-......)))))...)))......... ( -24.42) >DroEre_CAF1 102076 113 + 1 CAGCAACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU-----UUAU-AUUCUCUGCCGCA-AAAACCGAGAGCUCAGCACAAAAAUC ..((...(((((...(((((((.....)))))))......((((((((.......))))))))....)))-----))..-.(((((.......-......)))))....))......... ( -23.82) >DroYak_CAF1 104518 118 + 1 CAGCUACAAAACAAUGCUAGCCAAAACAGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUGUUUUCGUUUCGGUUUAU-UUUCUCGGCCGCA-AAAACCGAGAGCUCAGCACAAAAAUC ..(((.(.((((...((((((.......))))))......((((((((.......))))))))....)))).)......-.(((((((.....-....)))))))...)))......... ( -25.80) >consensus CAGCUACAAAACAAUGCUAGCCGAAAUGGUUAGCUAAAUUAUAACCAUCAUUUUAAUGGUUAUUUUCGUU_____UUAUUUUUCUCUGCCGCA_AAAACCGAGAGCUCAGCACAAAAAUC ..(((..........(((((((.....)))))))......((((((((.......))))))))..................(((((..............)))))...)))......... (-19.56 = -19.80 + 0.24)


| Location | 17,136,106 – 17,136,221 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136106 115 - 22407834 GAUUUUUGUGCUGAGCUCUCGGUUUUGUGCGGCAGAGAAAAAUAA-----AAAGAAAAUAACCAUUAAUAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCUG ..(((((.(((((.((..........)).))))).))))).((((-----((.....(((((((((.....)))))))))......((((.((.....)).))))....))))))..... ( -27.60) >DroSec_CAF1 104494 114 - 1 GAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCAGAGAAAAAUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCGG ..(((((.((((((((.....)))..-..))))).))))).((((-----(((((..(((((((((.....)))))))))......((((.((.....)).)))).)))))))))..... ( -30.90) >DroSim_CAF1 116687 114 - 1 GAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCAGAGAAAAAUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCUG ..(((((.((((((((.....)))..-..))))).))))).((((-----(((((..(((((((((.....)))))))))......((((.((.....)).)))).)))))))))..... ( -30.90) >DroEre_CAF1 102076 113 - 1 GAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCAGAGAAU-AUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUUGCUG ..(((((((((.((((.....)))).-.)).)))))))..-((((-----(((((..(((((((((.....)))))))))......((((.((.....)).)))).)))))))))..... ( -29.90) >DroYak_CAF1 104518 118 - 1 GAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCCGAGAAA-AUAAACCGAAACGAAAACAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACUGUUUUGGCUAGCAUUGUUUUGUAGCUG .........((((...((((((((..-...))))))))..-......((((((((....(((((((.....))))))).....((((((((.....))))))))..)))))))))))).. ( -32.90) >consensus GAUUUUUGUGCUGAGCUCUCGGUUUU_UGCGGCAGAGAAAAAUAA_____AACGAAAAUAACCAUUAAAAUGAUGGUUAUAAUUUAGCUAACCAUUUCGGCUAGCAUUGUUUUGUAGCUG ..(((((((((.((((.....))))...)).))))))).............(((((((((((((((.....)))))))))......((((.((.....)).))))....))))))..... (-24.50 = -24.94 + 0.44)

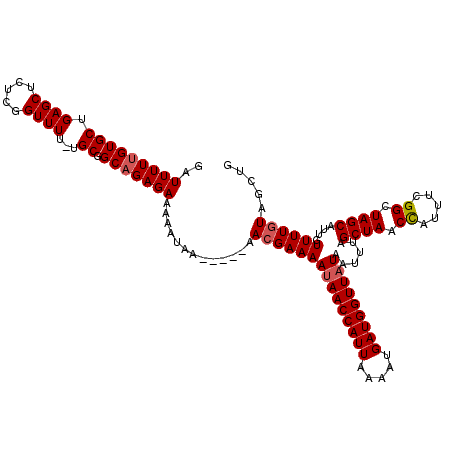
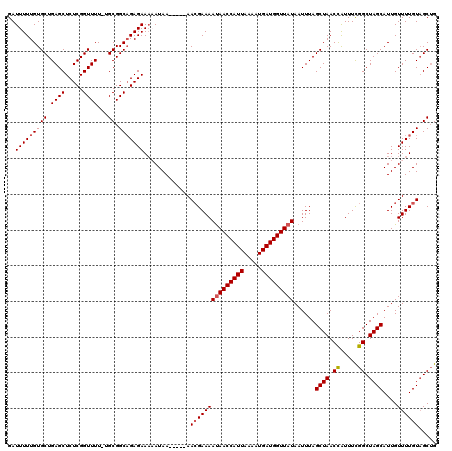
| Location | 17,136,146 – 17,136,261 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17136146 115 - 22407834 UGUUAUUGCUGUUGGCCAUCUUGAGCAUCAUUUGUGCGGCGAUUUUUGUGCUGAGCUCUCGGUUUUGUGCGGCAGAGAAAAAUAA-----AAAGAAAAUAACCAUUAAUAUGAUGGUUAU .....(((((((.((((.....((((.(((...(..(((......)))..))))))))..))))....)))))))..........-----.......(((((((((.....))))))))) ( -29.30) >DroSec_CAF1 104534 114 - 1 UGUUAUUGCUGUUGGCCAUCUUGAGCAUCAUUUGUGCGGCGAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCAGAGAAAAAUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAU .....(((((((.((((.....((((.(((...(..(((......)))..))))))))..))))..-.)))))))..........-----.......(((((((((.....))))))))) ( -29.90) >DroSim_CAF1 116727 114 - 1 UGUUAUUGCUGUUGGCCAUCUUGAGCAUCAUUUGUGCGGCGAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCAGAGAAAAAUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAU .....(((((((.((((.....((((.(((...(..(((......)))..))))))))..))))..-.)))))))..........-----.......(((((((((.....))))))))) ( -29.90) >DroEre_CAF1 102116 113 - 1 UGUUAUUGCUGUUGGCCAUCUUGAGCAUCAUUUGUUCGGCGAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCAGAGAAU-AUAA-----AACGAAAAUAACCAUUAAAAUGAUGGUUAU .....(((((((.((((....((.....))...((((((((.......))))))))....))))..-.))))))).....-....-----.......(((((((((.....))))))))) ( -30.00) >DroYak_CAF1 104558 118 - 1 UGUUAUUGCUGUUGGCCAUCUUGAGCAUCAUUUGCUCGGCGAUUUUUGUGCUGAGCUCUCGGUUUU-UGCGGCCGAGAAA-AUAAACCGAAACGAAAACAACCAUUAAAAUGAUGGUUAU .(((.(((...((((..................((((((((.......))))))))((((((((..-...))))))))..-.....))))..))).)))(((((((.....))))))).. ( -34.80) >consensus UGUUAUUGCUGUUGGCCAUCUUGAGCAUCAUUUGUGCGGCGAUUUUUGUGCUGAGCUCUCGGUUUU_UGCGGCAGAGAAAAAUAA_____AACGAAAAUAACCAUUAAAAUGAUGGUUAU .....(((((((.((((.....((((.(((...((((((......)))))))))))))..))))....)))))))......................(((((((((.....))))))))) (-26.74 = -27.38 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:25 2006