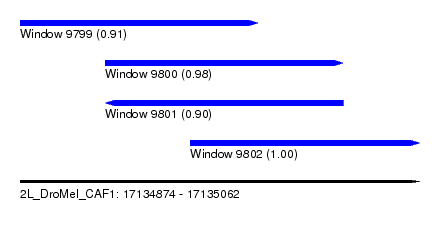
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,134,874 – 17,135,062 |
| Length | 188 |
| Max. P | 0.999904 |

| Location | 17,134,874 – 17,134,986 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -23.49 |
| Energy contribution | -25.17 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134874 112 + 22407834 GGAAGAAAGCACUUGAUUAGGAGAAAGAGCAGGCAACCGAAAGUAAAGGCUUGGGACUUUCGGACGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAU ........((((((..((.(..(((((..(((((...(....).....)))))...)))))...).))..))))))(((((((.(.((.......)).).)))))--------))..... ( -34.10) >DroSec_CAF1 103285 112 + 1 GGAAGAAAGCACUUGAUUAGGAGAAAGAGCAGGCAAGCGAAAGGAAAGGCUUGGGACUUCCGGGCGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAU (((((...((.(((..........))).))...(((((..........)))))...)))))............((((((((((.(.((.......)).).)))))--------))))).. ( -34.70) >DroSim_CAF1 115478 112 + 1 GGAAGAAAGCACUUGAUUAGGAGAAAGAGCAGGCAAGCGAAAGUAAAGGCUUGGGACUUCCGGACGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAU (((((......(((......)))......(((((..((....))....)))))...)))))............((((((((((.(.((.......)).).)))))--------))))).. ( -35.90) >DroEre_CAF1 100774 101 + 1 GGAUGAAAGCACUUGAUUAGCAGUAAGAGCAGGCAAGCGAAAGUAAAGGCUUCGGACUUCCGGAC-----------GGGCGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAU ........((.((((........)))).))..((..((....))....))(((((....))))).-----------.((.((((((((.........))))))))--------))..... ( -26.80) >DroYak_CAF1 103164 120 + 1 GGUAAAAAGCACUUGAUUAGGAGAAAGAGCAGGCAAGCGAAAGUAAAGGCUUGGGACUUCCGGACAGAGAAAGUGUGGCUGUGUGAGUAAAUAAGACGCUCACACCCACACACCCACAAU (((........(((.....((((......(((((..((....))....)))))....)))).....)))...((((((..((((((((.........)))))))))))))))))...... ( -36.50) >consensus GGAAGAAAGCACUUGAUUAGGAGAAAGAGCAGGCAAGCGAAAGUAAAGGCUUGGGACUUCCGGACGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC________CCACAAU (((((...((.(((..........))).))((((..((....))....))))....)))))((.........((((((((((.(.........).))))))))))........))..... (-23.49 = -25.17 + 1.68)


| Location | 17,134,914 – 17,135,026 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -31.11 |
| Energy contribution | -31.83 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134914 112 + 22407834 AAGUAAAGGCUUGGGACUUUCGGACGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCG .......((((((.(...(((((.(((((.((.((((((((((.(.((.......)).).)))))--------))))).))....))))).)))))...).))))))(((......))). ( -44.50) >DroSec_CAF1 103325 112 + 1 AAGGAAAGGCUUGGGACUUCCGGGCGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCG .......((((((.((((..(((.(((((.((.((((((((((.(.((.......)).).)))))--------))))).))....))))).)))))))...))))))(((......))). ( -45.20) >DroSim_CAF1 115518 112 + 1 AAGUAAAGGCUUGGGACUUCCGGACGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCG .......((((((.((((..(((.(((((.((.((((((((((.(.((.......)).).)))))--------))))).))....))))).)))))))...))))))(((......))). ( -42.70) >DroEre_CAF1 100814 101 + 1 AAGUAAAGGCUUCGGACUUCCGGAC-----------GGGCGUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCCUCGGCUGAGUCCUGUAAGCCGCUCACCAAAGCG .......((((((((....))((((-----------.((.((((((((.........))))))))--------))........(((....)))..)))).).)))))(((......))). ( -35.40) >DroYak_CAF1 103204 120 + 1 AAGUAAAGGCUUGGGACUUCCGGACAGAGAAAGUGUGGCUGUGUGAGUAAAUAAGACGCUCACACCCACACACCCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCG .......((((((((..((((.....).))).((((((..((((((((.........)))))))))))))).)))(((..(..(((....)))..)...))))))))(((......))). ( -42.60) >consensus AAGUAAAGGCUUGGGACUUCCGGACGGAGAGAGUGUGGGUGUGUGAGUAAAUAAGACGCUCACAC________CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCG .......((((((.((((((((((.........(((((..((((((((.........))))))))........)))))........)))))..)))))...))))))(((......))). (-31.11 = -31.83 + 0.72)

| Location | 17,134,914 – 17,135,026 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -28.34 |
| Energy contribution | -29.26 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134914 112 - 22407834 CGCUUUGGUGAGCGGCUUACACGACUCAGCCGAAGCUUUGAUUGUGG--------GUGUGAGCGUCUUAUUUACUCACACACCCACACUCUCUCCGUCCGAAAGUCCCAAGCCUUUACUU .(((((((((((((.......)).)))).)))))))...((.(((((--------(((((.(.((.......)).).)))))))))).)).....(..(....)..)............. ( -38.50) >DroSec_CAF1 103325 112 - 1 CGCUUUGGUGAGCGGCUUACACGACUCAGCCGAAGCUUUGAUUGUGG--------GUGUGAGCGUCUUAUUUACUCACACACCCACACUCUCUCCGCCCGGAAGUCCCAAGCCUUUCCUU .(((((((((((((.......)).)))).)))))))...((.(((((--------(((((.(.((.......)).).)))))))))).)).........(((((.........))))).. ( -39.40) >DroSim_CAF1 115518 112 - 1 CGCUUUGGUGAGCGGCUUACACGACUCAGCCGAAGCUUUGAUUGUGG--------GUGUGAGCGUCUUAUUUACUCACACACCCACACUCUCUCCGUCCGGAAGUCCCAAGCCUUUACUU .(((((((((((((.......)).)))).)))))))...((.(((((--------(((((.(.((.......)).).)))))))))).))..(((....))).................. ( -39.30) >DroEre_CAF1 100814 101 - 1 CGCUUUGGUGAGCGGCUUACAGGACUCAGCCGAGGCUUUGAUUGUGG--------GUGUGAGCGUCUUAUUUACUCACACGCCC-----------GUCCGGAAGUCCGAAGCCUUUACUU .((((....))))(((((...(((((...(((.(((.........((--------(((((.(.((.......)).).)))))))-----------)))))).))))).)))))....... ( -36.80) >DroYak_CAF1 103204 120 - 1 CGCUUUGGUGAGCGGCUUACACGACUCAGCCGAAGCUUUGAUUGUGGGUGUGUGGGUGUGAGCGUCUUAUUUACUCACACAGCCACACUUUCUCUGUCCGGAAGUCCCAAGCCUUUACUU .(((((((((((((.......)).)))).)))))))((((...((((.((((((((((((((...)))))..))))))))).))))((((((.......))))))..))))......... ( -44.40) >consensus CGCUUUGGUGAGCGGCUUACACGACUCAGCCGAAGCUUUGAUUGUGG________GUGUGAGCGUCUUAUUUACUCACACACCCACACUCUCUCCGUCCGGAAGUCCCAAGCCUUUACUU .(((((((((((((.......)).)))).)))))))...((.(((((........(((((((...........)))))))..))))).)).....(..(....)..)............. (-28.34 = -29.26 + 0.92)

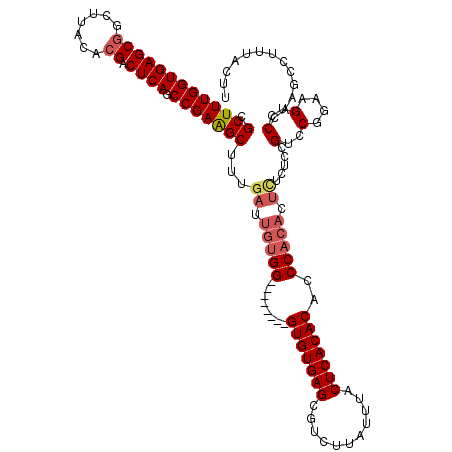

| Location | 17,134,954 – 17,135,062 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134954 108 + 22407834 GUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCGUAAAGACAAUUCAAUCGAUGGCAAAUACCCAUACAA---- ((((((((.........))))))))--------...........((((((..(((((.((.(....((((......))))...).)).)))))..))).)))..............---- ( -27.40) >DroSec_CAF1 103365 108 + 1 GUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCGUAAAGACAAUUCAAUCGAUGGCAAAUACCCAUACAA---- ((((((((.........))))))))--------...........((((((..(((((.((.(....((((......))))...).)).)))))..))).)))..............---- ( -27.40) >DroSim_CAF1 115558 108 + 1 GUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCGUAAAGACAAUUCAAUCGAUGGCAAAUACCCAUACAA---- ((((((((.........))))))))--------...........((((((..(((((.((.(....((((......))))...).)).)))))..))).)))..............---- ( -27.40) >DroEre_CAF1 100843 112 + 1 GUGUGAGUAAAUAAGACGCUCACAC--------CCACAAUCAAAGCCUCGGCUGAGUCCUGUAAGCCGCUCACCAAAGCGUAAAGACAAUUCAAUCGAUGGCGAAUACUCAUACAAUCGC (((((((((.....(.....)....--------...........((((((..(((((..(((....((((......)))).....))))))))..))).)))...)))))))))...... ( -30.50) >DroYak_CAF1 103244 120 + 1 GUGUGAGUAAAUAAGACGCUCACACCCACACACCCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCGUAAAGACAAUUCAAUCGAUGGCGAAUACUCAUACAAUCGC (((((((((......(((((.......................(((....)))((((.((....)).)))).....))))).............(((....))).)))))))))...... ( -30.00) >consensus GUGUGAGUAAAUAAGACGCUCACAC________CCACAAUCAAAGCUUCGGCUGAGUCGUGUAAGCCGCUCACCAAAGCGUAAAGACAAUUCAAUCGAUGGCAAAUACCCAUACAA____ ((((((((.........))))))))...................((((((..(((((..(((....((((......)))).....))))))))..))).))).................. (-27.36 = -27.20 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:19 2006