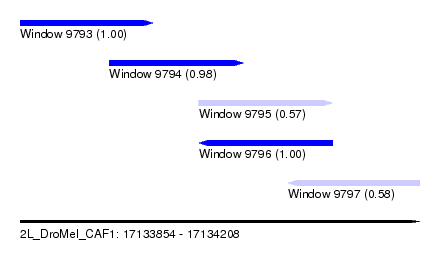
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,133,854 – 17,134,208 |
| Length | 354 |
| Max. P | 0.999201 |

| Location | 17,133,854 – 17,133,972 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -28.09 |
| Energy contribution | -28.01 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17133854 118 + 22407834 ACCUCUGUCUUCUCAAUUUUCCACCCAACUGAUGGGGAACGUGUAAAGGUUUAAAGUGUGGGCAACUGGAGGGA-AAAAGU-AGACAGAAAAACGGAAAAGCGCAAACAUUGCGCAGCAA ...(((((((.((...((((((..((.(((......((((.(....).))))..)))..((....))))..)))-))))).-)))))))...........(((((.....)))))..... ( -32.50) >DroSec_CAF1 102321 118 + 1 ACCUCUGUCUUCUCAAUUUUCCACCCAUCUUAUGGGGCACGUGCAAAGGUUCAAAGUGUAGGCAACUGGAGGAA-AAAUGU-AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAA ...(((((((...((.((((((.(((((...)))))((((..((....)).....))))((....))...))))-)).)).-)))))))...........(((((.....)))))..... ( -35.60) >DroSim_CAF1 114510 112 + 1 ACCUCUGUCUUCUCAAUUUUCCACCCAUCUUAUGGGGCACGUGCAAAGGUUCAAAGUGU-------UGGAGGAAAAAAAGU-AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAA ...(((((((.((...((((((.(((((...)))))((((..((....)).....))))-------....))))))..)).-)))))))...........(((((.....)))))..... ( -32.50) >DroEre_CAF1 99883 119 + 1 ACCUCUGUCUUCUUAAUUUUCCACCCAUCACUUGGGGCACGUGCAGAUGCUCAAAGUGUAGGCAACUGAAGGAA-CAAAAUAAGACAGAAAAACAGAGAAGCGCAAACAUUGCGCAGCAA ...((((((((.......((((......(((((.((((((.....).))))).)))))(((....)))..))))-......))))))))...........(((((.....)))))..... ( -35.62) >DroYak_CAF1 102227 118 + 1 ACCUCUGUCUUCUCAAUUUUCCACCCAUCACUUGGGGCACGUGCAAAGGUUCAAAGUGUAGGCAACUGAAGAAA-AAAAAC-AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAA ...(((((((......(((((..((((.....))))((((..((....)).....))))((....))...))))-).....-)))))))...........(((((.....)))))..... ( -29.10) >consensus ACCUCUGUCUUCUCAAUUUUCCACCCAUCUCAUGGGGCACGUGCAAAGGUUCAAAGUGUAGGCAACUGGAGGAA_AAAAGU_AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAA ...(((((((........((((.((((.....))))((((..((....)).....))))...........))))........)))))))...........(((((.....)))))..... (-28.09 = -28.01 + -0.08)


| Location | 17,133,933 – 17,134,052 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.36 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17133933 119 + 22407834 U-AGACAGAAAAACGGAAAAGCGCAAACAUUGCGCAGCAAACAAAGUCAGUGGCCAUUUGCUUGUUCUGCUCCCAACUCCUGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGU .-..((((......(((...(((((.....)))))(((((((((.((.(((....))).))))))).))))......)))(((((......))))).....))))............... ( -25.70) >DroSec_CAF1 102400 119 + 1 U-AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAAACAAAGUCAGUGGCCAUUUGCUUGUUCUGCUCCCAACUCCUGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGU .-..((((............(((((.....))))).....((((.((...((((((((.(((.(((........)))....)))....))))))))..)).))))...........)))) ( -25.60) >DroSim_CAF1 114583 119 + 1 U-AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAAACAAAGUCAGUGGCCAUUUGCUUGUUCUGCUCCCAACUCCUGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGU .-..((((............(((((.....))))).....((((.((...((((((((.(((.(((........)))....)))....))))))))..)).))))...........)))) ( -25.60) >DroEre_CAF1 99962 120 + 1 UAAGACAGAAAAACAGAGAAGCGCAAACAUUGCGCAGCAAACAAAGUCAGUGGCCAUUUGCCUGUUUUGCUCCCAACUCCUGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGU ....((((............(((((.....))))).....((((.((...((((((((.(((.(..(((....)))..)..)))....))))))))..)).))))...........)))) ( -27.80) >DroYak_CAF1 102306 119 + 1 C-AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAAACAAAGUCAGUGGCCAUUUGCCUGUUCUGCUCCCAACUCCUGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGU .-..((((............(((((.....))))).....((((.((...((((((((.(((.(((........)))....)))....))))))))..)).))))...........)))) ( -27.50) >consensus U_AGACAGAAAAACAGAAAAGCGCAAACAUUGCGCAGCAAACAAAGUCAGUGGCCAUUUGCUUGUUCUGCUCCCAACUCCUGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGU ....((((............(((((.....))))).....((((.((...((((((((.(((.(((........)))....)))....))))))))..)).))))...........)))) (-26.60 = -26.36 + -0.24)

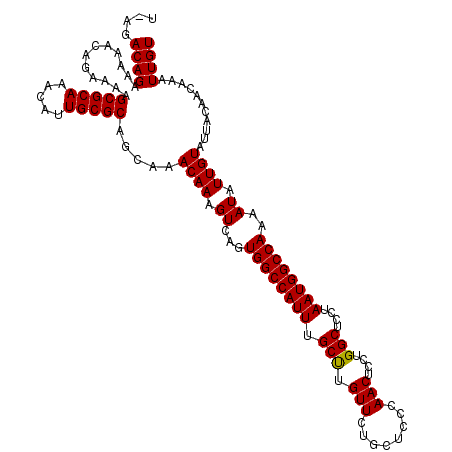
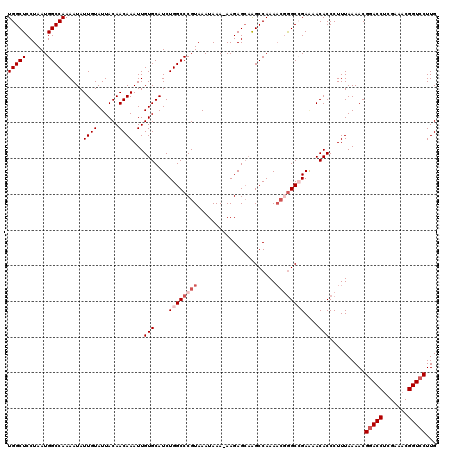
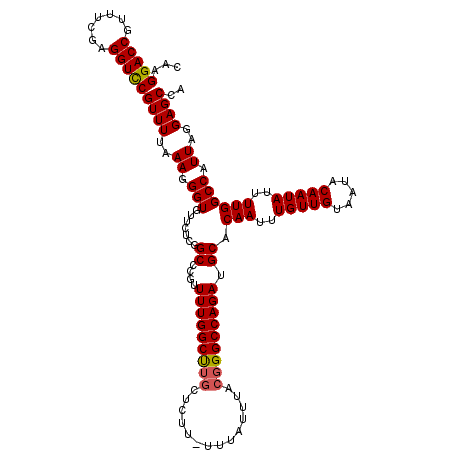
| Location | 17,134,012 – 17,134,131 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -21.25 |
| Energy contribution | -22.85 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134012 119 + 22407834 UGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGUGCAUCUGGCCCGUAAAUAAA-AAGAGCAAGCCAAAACGGGCCGAAAACACCCUUUAAAACGGACCUCGAAACGGUCCUUG (((((......))))).....((((......))))...(((....((((((((.......-..............))))))))....)))..........(((((.......)))))... ( -29.50) >DroSec_CAF1 102479 119 + 1 UGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGUGCAUCUGGCCCGUAAAUAAA-AAGAGCAAGCCAAAACGGGCCGAGAACACCCUUUAAAACGGACCUCGAAACGGUCCUUG (((((......))))).....((((......))))...(((..((((((((((.......-..............))))))).))).)))..........(((((.......)))))... ( -31.00) >DroSim_CAF1 114662 114 + 1 UGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGUGCAUCUGGCCCGUAAAUAAA-AAGAGCAAGCCAAAACGGGCCGAGAACACCCUUUAAAACGGACCUC-----GGUCCUUG (((((......))))).....((((......))))...(((..((((((((((.......-..............))))))).))).)))..........((((...-----.))))... ( -29.50) >DroEre_CAF1 100042 119 + 1 UGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGUGCAUCUGGCCCGUAAAUAAAAAAGAGAAGGCCAAAAC-AGCAGAAAACACCCUUUAAAACGGACCUCGAAACGGUCCUUG (((((......))))).....((((......)))).((.(((...(((((..................)))))....-.))).))...............(((((.......)))))... ( -24.27) >DroYak_CAF1 102385 117 + 1 UGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGUGCAUCUGGCCCGUAAAUAAAAAAGAGCUGGCCAAACA-GGCAGA--ACACCCUUUAAAACGAACCUCGAAACGGUCCUUG (((((......)))))......(((((...........)))))...((.((((..........(..(((.((.....-))))).--.)...........((.....))..)))).))... ( -20.50) >consensus UGGCUCCUAAUGGCCAAAAUAUUGUAUUACAACAAAUUGUGCAUCUGGCCCGUAAAUAAA_AAGAGCAAGCCAAAACGGGCCGAAAACACCCUUUAAAACGGACCUCGAAACGGUCCUUG (((((......))))).....((((......))))...(((....((((((((......................))))))))....)))..........(((((.......)))))... (-21.25 = -22.85 + 1.60)


| Location | 17,134,012 – 17,134,131 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -31.04 |
| Energy contribution | -31.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134012 119 - 22407834 CAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUUUCGGCCCGUUUUGGCUUGCUCUU-UUUAUUUACGGGCCAGAUGCACAAUUUGUUGUAAUACAAUAUUUUGGCCAUUAGGAGCCA ........(((((..(((((((..(((((((.((....((((......)))).)).)))-))))...))))))).((((..(((..(((((.....)))))..)))..)))).))))).. ( -37.30) >DroSec_CAF1 102479 119 - 1 CAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUCUCGGCCCGUUUUGGCUUGCUCUU-UUUAUUUACGGGCCAGAUGCACAAUUUGUUGUAAUACAAUAUUUUGGCCAUUAGGAGCCA ........(((((..(((((((..(((((((.((....((((......)))).)).)))-))))...))))))).((((..(((..(((((.....)))))..)))..)))).))))).. ( -37.30) >DroSim_CAF1 114662 114 - 1 CAAGGACC-----GAGGUCCGUUUUAAAGGGUGUUCUCGGCCCGUUUUGGCUUGCUCUU-UUUAUUUACGGGCCAGAUGCACAAUUUGUUGUAAUACAAUAUUUUGGCCAUUAGGAGCCA ...((.((-----..(((((((..(((((((.((....((((......)))).)).)))-))))...))))))).((((..(((..(((((.....)))))..)))..)))).))..)). ( -37.20) >DroEre_CAF1 100042 119 - 1 CAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUUUCUGCU-GUUUUGGCCUUCUCUUUUUUAUUUACGGGCCAGAUGCACAAUUUGUUGUAAUACAAUAUUUUGGCCAUUAGGAGCCA ...(((((.......)))))((((..((.(((......(((.-..((((((((................)))))))).)))(((..(((((.....)))))..)))))).))..)))).. ( -34.19) >DroYak_CAF1 102385 117 - 1 CAAGGACCGUUUCGAGGUUCGUUUUAAAGGGUGU--UCUGCC-UGUUUGGCCAGCUCUUUUUUAUUUACGGGCCAGAUGCACAAUUUGUUGUAAUACAAUAUUUUGGCCAUUAGGAGCCA ........(((((..(((((((...(((((((..--...(((-.....)))..))))))).......))))))).((((..(((..(((((.....)))))..)))..)))).))))).. ( -32.80) >consensus CAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUCUCGGCCCGUUUUGGCUUGCUCUU_UUUAUUUACGGGCCAGAUGCACAAUUUGUUGUAAUACAAUAUUUUGGCCAUUAGGAGCCA ...(((((.......)))))((((..((.(((.......((....(((((((((..............))))))))).)).(((..(((((.....)))))..)))))).))..)))).. (-31.04 = -31.04 + -0.00)

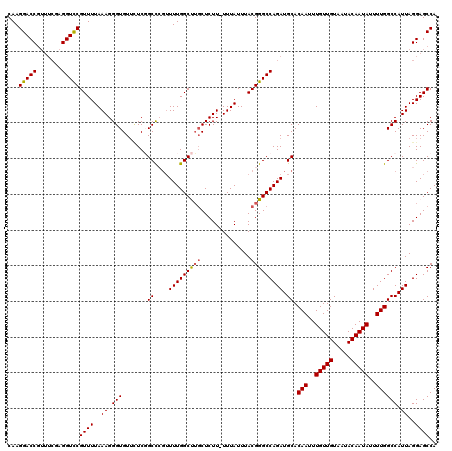
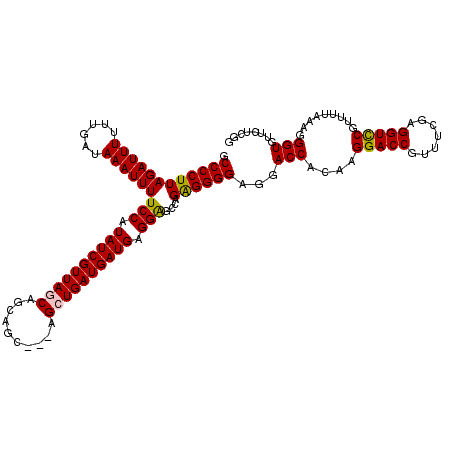
| Location | 17,134,091 – 17,134,208 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -36.89 |
| Consensus MFE | -32.74 |
| Energy contribution | -32.66 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17134091 117 - 22407834 GCCCCUUAGAUUUUUUGAUAAAUUUUCCAUAUCGUUAGCAGCAGC---AGCUGAUGAUGAGGAGCCAGAGGGGAGGACCACAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUUUCGG .((((((((((((......))))))(((.((((((((((......---.)))))))))).)))....))))))((.(((....(((((.......))))).........))).))..... ( -37.22) >DroSec_CAF1 102558 120 - 1 GCCCCUUAGAUUUUUUGAUAAAUUUUCCAUAUCGUUAGCAGCAGCGGCAGAUGAUGAUGAGGAGCCAGAGGGGAGGACCACAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUCUCGG ..(((((((((((......))))))(((.((((((((...((....))...)))))))).)))....)))))((((((.((..(((((.......)))))..........)))))))).. ( -35.50) >DroSim_CAF1 114741 115 - 1 GCCCCUUAGAUUUUUUGAUAAAUUUUCCAUAUCGUUAGCAGCAGCGGCAGAUGAUGAUGAGGGGCCAGAGGGGAGGACCACAAGGACC-----GAGGUCCGUUUUAAAGGGUGUUCUCGG (((((((((((((......))))).....(((((((.((.......)).))))))).)))))))).....(((((.(((....((((.-----...)))).........))).))))).. ( -37.32) >DroEre_CAF1 100121 117 - 1 GCCCCUUAGAUUUUUUGAUAAAUUUUCCAUAUCGUUAGCAGCAGC---AGCUGAUGAUGAGGAGCCAGGGGGGAGGACCACAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUUUCUG .(((((.((((((......))))))(((.((((((((((......---.)))))))))).)))...)))))(((((((.((..(((((.......)))))..........))))))))). ( -38.80) >DroYak_CAF1 102464 115 - 1 GCCCCUUAGAUUUUUUGAUAAAUUUUCCAUAUCGUUAGCAGCAGC---AGCUGAUGAUGAGGAGCCAGAGGGGAGGACCACAAGGACCGUUUCGAGGUUCGUUUUAAAGGGUGU--UCUG .((((((((((((......))))))(((.((((((((((......---.)))))))))).)))....)))))).(((.(((..(((((.......)))))..........))).--))). ( -35.60) >consensus GCCCCUUAGAUUUUUUGAUAAAUUUUCCAUAUCGUUAGCAGCAGC___AGCUGAUGAUGAGGAGCCAGAGGGGAGGACCACAAGGACCGUUUCGAGGUCCGUUUUAAAGGGUGUUCUCGG .((((((((((((......))))))(((.((((((((((..........)))))))))).)))....))))))...(((....(((((.......))))).........)))........ (-32.74 = -32.66 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:14 2006