
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,131,198 – 17,131,394 |
| Length | 196 |
| Max. P | 0.980526 |

| Location | 17,131,198 – 17,131,314 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.02 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17131198 116 + 22407834 AAGAGCCAUCCUGGCAUCGGGAAAUCCAAAGGCGCCACC-CAC---CGCCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCG ..((((((((.((((...((((........((((.....-...---))))...)))).................((..((((.....))))..)).)))).).....)))))))...... ( -29.10) >DroSec_CAF1 99676 116 + 1 AAGAGCCAUCCUGGCAUCGGGAAAUCCAAAGGCGCCACC-CAC---CACCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCG ..(((((((((((....)))))........((((((...-...---....................(((...)))...((((.....)))).))).))).........))))))...... ( -27.60) >DroSim_CAF1 111603 116 + 1 AAGAGCCAUCCUGGCAUCGGGAAAUCCAAAGGCGCCACC-CAC---CACCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCG ..(((((((((((....)))))........((((((...-...---....................(((...)))...((((.....)))).))).))).........))))))...... ( -27.60) >DroEre_CAF1 97385 117 + 1 UAGAGCCAUCCUGGCAUCGGGAAAUCCAAAGACGCCACCGCAC---CAUCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCG ...........((((.((.((....))...)).)))).(((..---............................((..((((.....))))..))((((((......))))))....))) ( -29.00) >DroYak_CAF1 97782 120 + 1 UAGAGCCAUCCUGGCAUCGGGAAAUCCAAAGGCGCUACCGCACCACCAUCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCG ....(((.(((((....)))))........(((((....))...........((((......(((......)))......))))..)))...)))((((((......))))))....... ( -29.80) >consensus AAGAGCCAUCCUGGCAUCGGGAAAUCCAAAGGCGCCACC_CAC___CACCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCG ..(((((((..((((.((.((....))...)).))))..............................(((((..((..((((.....))))..))....)))))...)))))))...... (-27.34 = -27.02 + -0.32)

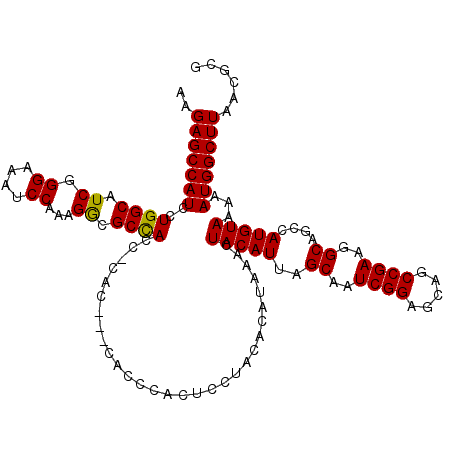
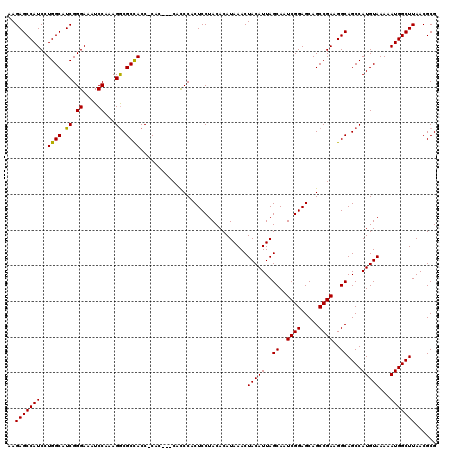
| Location | 17,131,237 – 17,131,354 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.14 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17131237 117 + 22407834 CAC---CGCCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAU ...---(((.........................((..((((.....))))..))((((((......))))))....))).................(((((....)))))......... ( -21.10) >DroSec_CAF1 99715 117 + 1 CAC---CACCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAU ...---......((((......(((......)))......)))).....((((((((((((......))))))........................(((((....)))))))))))... ( -19.50) >DroSim_CAF1 111642 117 + 1 CAC---CACCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAU ...---......((((......(((......)))......)))).....((((((((((((......))))))........................(((((....)))))))))))... ( -19.50) >DroEre_CAF1 97425 117 + 1 CAC---CAUCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAU .((---(((.........................((..((((.....))))..))((((((......))))))...................................)))))....... ( -19.90) >DroYak_CAF1 97822 120 + 1 CACCACCAUCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAU ....(((((.........................((..((((.....))))..))((((((......))))))...................................)))))....... ( -19.90) >consensus CAC___CACCCACUCCUACACAUAAACUACAUUAGCAAUCGGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAU ............((((......(((......)))......)))).....((((((((((((......))))))........................(((((....)))))))))))... (-19.50 = -19.50 + 0.00)

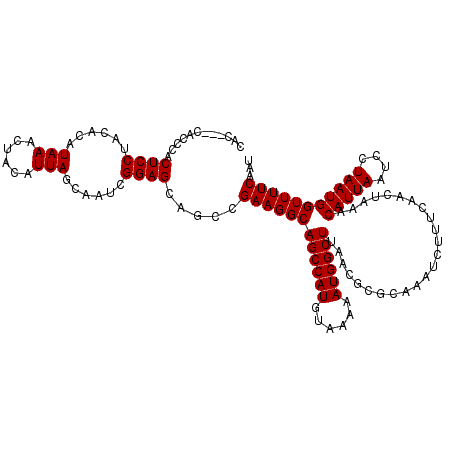
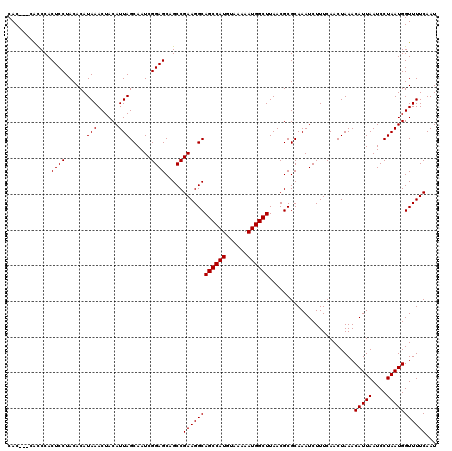
| Location | 17,131,237 – 17,131,354 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.14 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17131237 117 - 22407834 AUUGAAAACCAUUAGGAUUAAUGUUUAGUUGAAAGAUUUGCGCGUUAAGCCAUUUUUACAUGGCUGCCUUCGGCUGCUCCGAUUGCUAAUGUAGUUUAUGUGUAGGAGUGGGCG---GUG .......(((.......(((((((..((((....))))...))))))).(((((((((((((((.(((...))).)))..((((((....))))))...))))))))))))..)---)). ( -33.00) >DroSec_CAF1 99715 117 - 1 AUUGAAAACCAUUAGGAUUAAUGUUUAGUUGAAAGAUUUGCGCGUUAAGCCAUUUUUACAUGGCUGCCUUCGGCUGCUCCGAUUGCUAAUGUAGUUUAUGUGUAGGAGUGGGUG---GUG .......(((((.....(((((((..((((....))))...))))))).(((((((((((((((.(((...))).)))..((((((....))))))...)))))))))))))))---)). ( -34.50) >DroSim_CAF1 111642 117 - 1 AUUGAAAACCAUUAGGAUUAAUGUUUAGUUGAAAGAUUUGCGCGUUAAGCCAUUUUUACAUGGCUGCCUUCGGCUGCUCCGAUUGCUAAUGUAGUUUAUGUGUAGGAGUGGGUG---GUG .......(((((.....(((((((..((((....))))...))))))).(((((((((((((((.(((...))).)))..((((((....))))))...)))))))))))))))---)). ( -34.50) >DroEre_CAF1 97425 117 - 1 AUUGAAAACCAUUAGGAUUAAUGUUUAGUUGAAAGAUUUGCGCGUUAAGCCAUUUUUACAUGGCUGCCUUCGGCUGCUCCGAUUGCUAAUGUAGUUUAUGUGUAGGAGUGGAUG---GUG .......(((((.....(((((((..((((....))))...))))))).(((((((((((((((.(((...))).)))..((((((....))))))...)))))))))))))))---)). ( -34.00) >DroYak_CAF1 97822 120 - 1 AUUGAAAACCAUUAGGAUUAAUGUUUAGUUGAAAGAUUUGCGCGUUAAGCCAUUUUUACAUGGCUGCCUUCGGCUGCUCCGAUUGCUAAUGUAGUUUAUGUGUAGGAGUGGAUGGUGGUG .......(((((((...(((((((..((((....))))...))))))).(((((((((((((((.(((...))).)))..((((((....))))))...)))))))))))).))))))). ( -36.40) >consensus AUUGAAAACCAUUAGGAUUAAUGUUUAGUUGAAAGAUUUGCGCGUUAAGCCAUUUUUACAUGGCUGCCUUCGGCUGCUCCGAUUGCUAAUGUAGUUUAUGUGUAGGAGUGGGUG___GUG ........((....)).(((((((..((((....))))...))))))).(((((((((((((((.(((...))).)))..((((((....))))))...))))))))))))......... (-30.32 = -30.32 + 0.00)

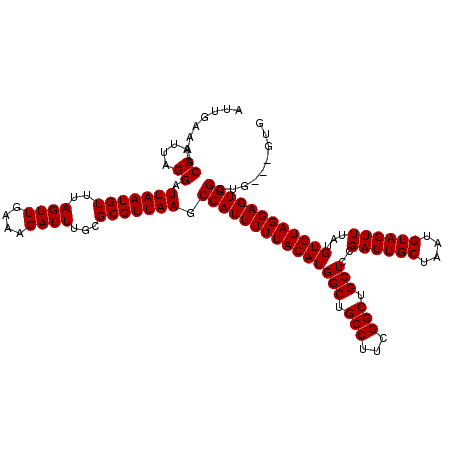
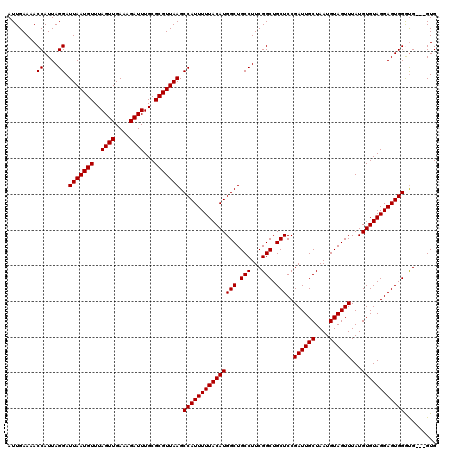
| Location | 17,131,274 – 17,131,394 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -33.53 |
| Energy contribution | -33.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17131274 120 + 22407834 GGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAUUUUGUUGCGCCUUCUGUGGCAAGUCAAGUUCGCGUCUCCU (((((.(((((((((((((((......))))))......((((..............(((((....))))).............))))))))))...)))..((.......))).)))). ( -33.53) >DroSec_CAF1 99752 120 + 1 GGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAUUUUGUUGCGCCUUCUGUGGCAAGUCAAGUUCGCGUCUCCU (((((.(((((((((((((((......))))))......((((..............(((((....))))).............))))))))))...)))..((.......))).)))). ( -33.53) >DroSim_CAF1 111679 120 + 1 GGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAUUUUGUUGCGCCUUCUGUGGCAAGUCAAGUUCGCGUCUCCU (((((.(((((((((((((((......))))))......((((..............(((((....))))).............))))))))))...)))..((.......))).)))). ( -33.53) >DroEre_CAF1 97462 120 + 1 GGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAUUUUGUUGCGCCUUCUGUGGCAAGUCAAGUUCGCGUCUCCU (((((.(((((((((((((((......))))))......((((..............(((((....))))).............))))))))))...)))..((.......))).)))). ( -33.53) >DroYak_CAF1 97862 120 + 1 GGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAUUUUGUUGCGCCUUCUGUGGCAAGUCAAGUUCGCGUCUCCU (((((.(((((((((((((((......))))))......((((..............(((((....))))).............))))))))))...)))..((.......))).)))). ( -33.53) >consensus GGAGCAGCCGAAGGCAGCCAUGUAAAAAUGGCUUAACGCGCAAAUCUUUCAACUAAACAUUAAUCCUAAUGGUUUUCAAUUUUGUUGCGCCUUCUGUGGCAAGUCAAGUUCGCGUCUCCU (((((.(((((((((((((((......))))))......((((..............(((((....))))).............))))))))))...)))..((.......))).)))). (-33.53 = -33.53 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:06 2006