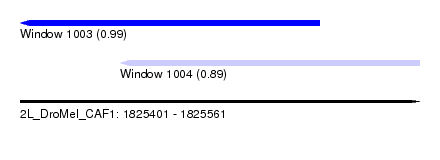
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,825,401 – 1,825,561 |
| Length | 160 |
| Max. P | 0.992370 |

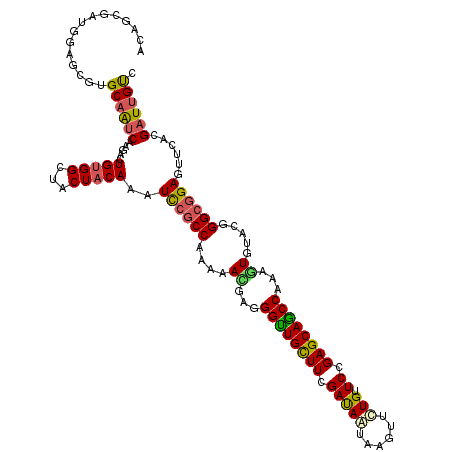
| Location | 1,825,401 – 1,825,521 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -42.43 |
| Consensus MFE | -35.32 |
| Energy contribution | -34.44 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1825401 120 - 22407834 AUUAGCCAAAAACGAGGGUUGCUUCGAUAAUAAGUUCUGUUCCGAGCAGCCAAAAUGUACGGGCGGAGUUCAUGAUUGUCGCUUUGUGGAGUCGAACAUGAAGAUUUGUCAGGCGGAAAA ....(((.........((((((((.((.............)).))))))))..........((((((.((((((....((((((....))).))).))))))..)))))).)))...... ( -33.62) >DroSec_CAF1 45462 120 - 1 CUCCGCCAAAAACGAGGGUUGCUUCGACAAUAAGUUUUGUUCCGAGCAACCAAAGUGUACGGGCGGAGUUCACGAUUGUCGCUUUGUGGAGUCAAGUAUGCAGAUUUGUCAGGCGGAAAA .((((((....(((((((((((((.(((((......))).)).))))))))...((((((.....((.(((((((........))))))).))..))))))...)))))..))))))... ( -44.00) >DroSim_CAF1 23171 120 - 1 CUCCGCCAAAAACGAGGGUUGCUUCGAUAAUAAGUUCUGUUCCGAGCAACCAAAGUGUACGGGCGGAGUUCACGAUUGUCGCUUUGUGGAGUCAAGUAUGCAGAUUUGCCAGGCGGAAAA .((((((.........((((((((.((.............)).))))))))...((((((.....((.(((((((........))))))).))..))))))..........))))))... ( -40.72) >DroEre_CAF1 33935 120 - 1 AUCCGCCAGAAAUGAGGGCUGCUUCGAUAGUGAGUUCUGUUCCGAGCAGCCCAAGUGUUCGGGAGGAGUGCACGAUUGCCGCUUUGUGGAGUCGAAUAUGCAGAUUUGCCAGGCGGAAAA .((((((.(((((..(((((((((.(((((......))).)).)))))))))..)).)))((.(((..(((((((((.((((...)))))))))....))))..))).)).))))))... ( -50.20) >DroYak_CAF1 10328 120 - 1 GUCCGCCAGAAAUGAGGGCUGUUUCGAUAGUAAGUUCUGUUCCGAGCAGCCAAAAUGUUCGGGCGGAGUGCACGAUUGCCGCUUUGUGGAGUCGUCUAUGAAGAUUUGUCAGGCGGAAAA .((((((.(((((.......)))))(((((((...(((((.(((((((.......)))))))))))).)))((((((.((((...))))))))))...........)))).))))))... ( -43.60) >consensus AUCCGCCAAAAACGAGGGUUGCUUCGAUAAUAAGUUCUGUUCCGAGCAGCCAAAGUGUACGGGCGGAGUUCACGAUUGUCGCUUUGUGGAGUCGAAUAUGCAGAUUUGUCAGGCGGAAAA .((((((....((...((((((((.(((((......))).)).))))))))...)).....))))))...........((((((.(..((.((.........))))..).)))))).... (-35.32 = -34.44 + -0.88)

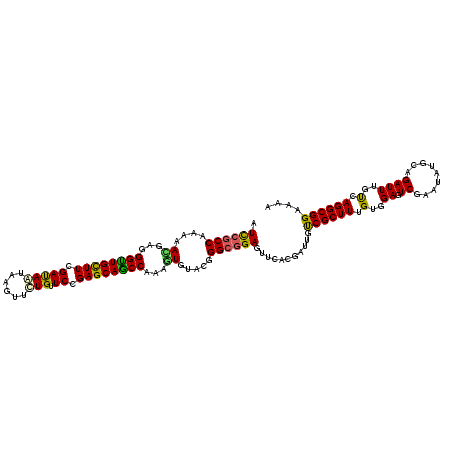
| Location | 1,825,441 – 1,825,561 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -32.18 |
| Energy contribution | -31.90 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1825441 120 - 22407834 ACAGCGAUGGAGCGUGCAAUCAGGAAUGUGGUUACUACAAAUUAGCCAAAAACGAGGGUUGCUUCGAUAAUAAGUUCUGUUCCGAGCAGCCAAAAUGUACGGGCGGAGUUCAUGAUUGUC ...((......))..(((((((.((((.((((((........))))))....(((((....))))).........(((((.(((.(((.......))).)))))))))))).))))))). ( -32.90) >DroSec_CAF1 45502 120 - 1 AUAGCGAUGGAGCCUGCAAUCAUGACUGUGGCUACUACAACUCCGCCAAAAACGAGGGUUGCUUCGACAAUAAGUUUUGUUCCGAGCAACCAAAGUGUACGGGCGGAGUUCACGAUUGUC ...(((((.(((((.(((........))))))).....(((((((((....((...((((((((.(((((......))).)).))))))))...)).....)))))))))..).))))). ( -45.30) >DroSim_CAF1 23211 120 - 1 AUAGCGAUGGAGCCUGCAAUCAUGACUGUGGCUACUACAACUCCGCCAAAAACGAGGGUUGCUUCGAUAAUAAGUUCUGUUCCGAGCAACCAAAGUGUACGGGCGGAGUUCACGAUUGUC ...(((((.(((((.(((........))))))).....(((((((((....((...((((((((.((.............)).))))))))...)).....)))))))))..).))))). ( -42.52) >DroEre_CAF1 33975 120 - 1 ACGACGAUGGAACGUGCCGCCGAGACUGUGGCUACUACAAAUCCGCCAGAAAUGAGGGCUGCUUCGAUAGUGAGUUCUGUUCCGAGCAGCCCAAGUGUUCGGGAGGAGUGCACGAUUGCC ............((((((((((......)))).........(((.((.(((((..(((((((((.(((((......))).)).)))))))))..)).))).)).)))).)))))...... ( -42.20) >DroYak_CAF1 10368 120 - 1 ACAGCGAUGGAGUGUGCAGUCGAGAUUGCGGCUACUACAAGUCCGCCAGAAAUGAGGGCUGUUUCGAUAGUAAGUUCUGUUCCGAGCAGCCAAAAUGUUCGGGCGGAGUGCACGAUUGCC ...(((((...(((..((((((......)))))........((((((.(((.....((((((((.(((((......))).)).))))))))......))).)))))))..))).))))). ( -47.30) >consensus ACAGCGAUGGAGCGUGCAAUCAAGACUGUGGCUACUACAAAUCCGCCAAAAACGAGGGUUGCUUCGAUAAUAAGUUCUGUUCCGAGCAGCCAAAGUGUACGGGCGGAGUUCACGAUUGUC ...............((((((.....(((((...)))))..((((((....((...((((((((.(((((......))).)).))))))))...)).....))))))......)))))). (-32.18 = -31.90 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:59 2006