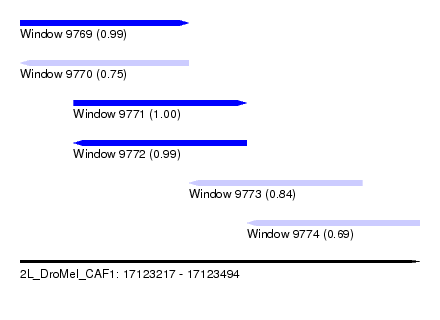
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,123,217 – 17,123,494 |
| Length | 277 |
| Max. P | 0.996773 |

| Location | 17,123,217 – 17,123,334 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -30.17 |
| Energy contribution | -32.42 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123217 117 + 22407834 GGAAAGCAGAAA---AAACACAACGGCGGCCAAAACAGAGUGUUGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCCACAUACGCCACGAUGUCUGGGCGUGUGUGCGAGUUCCG ((((..(.....---......................(((((((..((((((((((....))))))))))....)))))))((((((((((.((.....)))))))))))).)..)))). ( -42.10) >DroSec_CAF1 87035 100 + 1 GGAAAGCAGAAAAAAAAACACAACGGCGGCCAAAACAGAGUGAAGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCC--------------------GCGUGUGUGUGAGUUCCG ((((..((.........(((((...((((.........((((....((((((((((....))))))))))......))))))--------------------)).))))).))..)))). ( -26.80) >DroSim_CAF1 103100 119 + 1 GGAAAGCAGAAAA-AAAACACAACGGCGGCCAAAACAGAGUGAAGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGUUCCG ((((.........-..((((((...((.(((......).))...((((((.....))))))))..)))))).......(((((((((((((.((.....)))))))))))).))))))). ( -38.90) >DroEre_CAF1 89145 117 + 1 GGAAAGCAGAAA---AAACACAACGGCAGCCGAAGCAGAAUGUAGCAUACAUUGCGUAUGCGUAAUGUGUAACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGAUCCG (((.........---..(((((...((.((....))........((((((.....))))))))..)))))........(((((((((((((.((.....)))))))))))).))).))). ( -39.70) >DroYak_CAF1 88929 117 + 1 GGAAGGCAGAAA---AAACACAACGGCAGCCAAAACAGAGUGAAGCAUACAUUGCGUAUGCGUAAUGUGUAACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGUUCCG (((((((.....---.............)))...............((((((((((....))))))))))........(((((((((((((.((.....)))))))))))).))))))). ( -39.27) >consensus GGAAAGCAGAAA___AAACACAACGGCGGCCAAAACAGAGUGAAGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGUUCCG ((((............((((((..((...)).............((((((.....))))))....)))))).......(((((((((((((..........)))))))))).))))))). (-30.17 = -32.42 + 2.25)

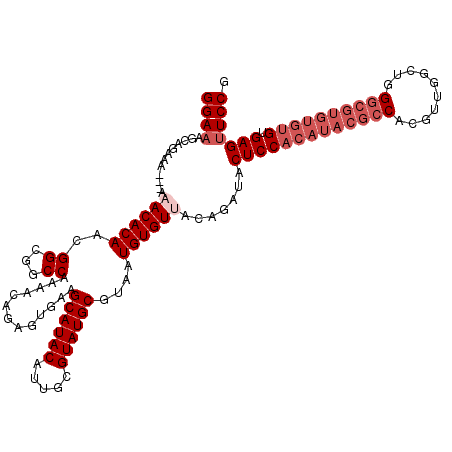
| Location | 17,123,217 – 17,123,334 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -25.23 |
| Energy contribution | -26.72 |
| Covariance contribution | 1.49 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123217 117 - 22407834 CGGAACUCGCACACACGCCCAGACAUCGUGGCGUAUGUGGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCAACACUCUGUUUUGGCCGCCGUUGUGUUU---UUUCUGCUUUCC ((((((((.((((.(((((..(....)..))))).)))))))).)))).((((((..((((((((.....)))))).............((...)))))))))).---............ ( -33.00) >DroSec_CAF1 87035 100 - 1 CGGAACUCACACACACGC--------------------GGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCUUCACUCUGUUUUGGCCGCCGUUGUGUUUUUUUUUCUGCUUUCC (((((...(((((.((((--------------------(((((....((((....))))((((((.....))))))...)))))))...((...)))))))))......)))))...... ( -22.70) >DroSim_CAF1 103100 119 - 1 CGGAACUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCUUCACUCUGUUUUGGCCGCCGUUGUGUUUU-UUUUCUGCUUUCC ((((((((.((((.((((((.......).))))).)))))))).)))).((((((..((((((((.....)))))).............((...))))))))))..-............. ( -32.40) >DroEre_CAF1 89145 117 - 1 CGGAUCUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUUACACAUUACGCAUACGCAAUGUAUGCUACAUUCUGCUUCGGCUGCCGUUGUGUUU---UUUCUGCUUUCC ((((....(((((.(((..(((((.((((.(((((((((..(((.....)))......))))))))).))))..((........))...))))).))))))))..---..))))...... ( -37.00) >DroYak_CAF1 88929 117 - 1 CGGAACUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUUACACAUUACGCAUACGCAAUGUAUGCUUCACUCUGUUUUGGCUGCCGUUGUGUUU---UUUCUGCCUUCC (((((...(((((.(((..((((((((((.(((((((((..(((.....)))......))))))))).))))..((........))..)))))).))))))))..---.)))))...... ( -35.80) >consensus CGGAACUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCUUCACUCUGUUUUGGCCGCCGUUGUGUUU___UUUCUGCUUUCC ((((((((.((((.(((((..........))))).)))))))).))))..(((((..((((((((.....))))))........((....))....)))))))................. (-25.23 = -26.72 + 1.49)

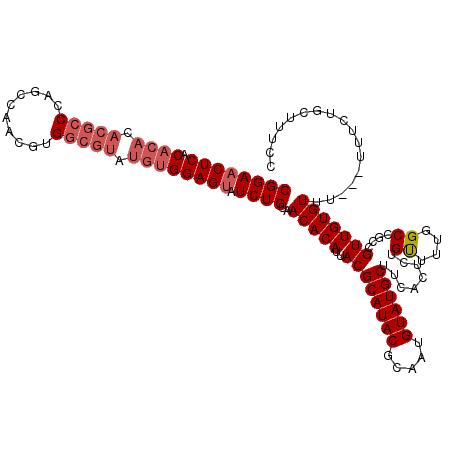
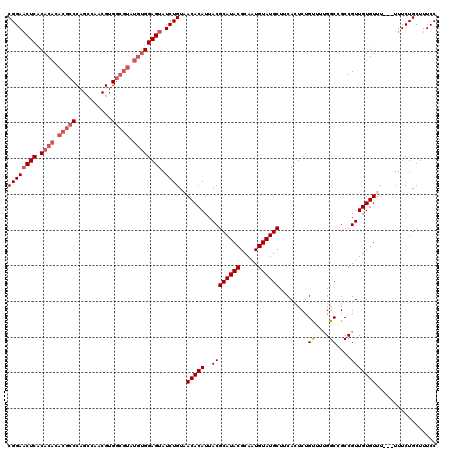
| Location | 17,123,254 – 17,123,374 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -31.77 |
| Energy contribution | -33.62 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123254 120 + 22407834 UGUUGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCCACAUACGCCACGAUGUCUGGGCGUGUGUGCGAGUUCCGUUUCCGCAAUUAAAAUGUUGAUUACAUGAAGUACGACUGC .(((((((((((((((....)))))))))).((.((.((((((((((((((.((.....)))))))))))).)))))).))....)))))......((((..(((.....)))))))... ( -41.50) >DroSec_CAF1 87075 100 + 1 UGAAGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCC--------------------GCGUGUGUGUGAGUUCCGUUUCCGCAAUUAAAAUGUUGAUUACAUGAAGUACGACUGC .(((((.(((((((((....)))))))))(((((.((((...--------------------..)))).))))).....)))))............((((..(((.....)))))))... ( -24.90) >DroSim_CAF1 103139 120 + 1 UGAAGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGUUCCGUUUCCGCAAUUAAAAUGUUGAUUACAUGAAGUACGACUGC .(((((((((((((((....))))))))))....(..((((((((((((((.((.....)))))))))))).))))..))))))............((((..(((.....)))))))... ( -40.70) >DroEre_CAF1 89182 120 + 1 UGUAGCAUACAUUGCGUAUGCGUAAUGUGUAACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGAUCCGUUUCCGCAAUUAAAAUGUUGAUUACAUGAAGUACGACUGC .(((((((((((((((....))))))))))(((.(((.(((((((((((((.((.....)))))))))))).)))))).)))...)).......((((.....))))....)))...... ( -40.90) >DroYak_CAF1 88966 120 + 1 UGAAGCAUACAUUGCGUAUGCGUAAUGUGUAACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGUUCCGUUUCCGCAAUUAAAAUGUUGAUUACAUGAAGUACGACUGC .(((((((((((((((....))))))))))....(..((((((((((((((.((.....)))))))))))).))))..))))))............((((..(((.....)))))))... ( -41.30) >consensus UGAAGCAUACAUUGCGUAUGCGUAAUGUGUUACAGAUACUCCACAUACGCCACGUUGGCUGGGCGUGUGUGUGAGUUCCGUUUCCGCAAUUAAAAUGUUGAUUACAUGAAGUACGACUGC ....((((((((((((....)))))))))).((.((.((((((((((((((..........)))))))))).)))))).))....)).........((((..(((.....)))))))... (-31.77 = -33.62 + 1.85)

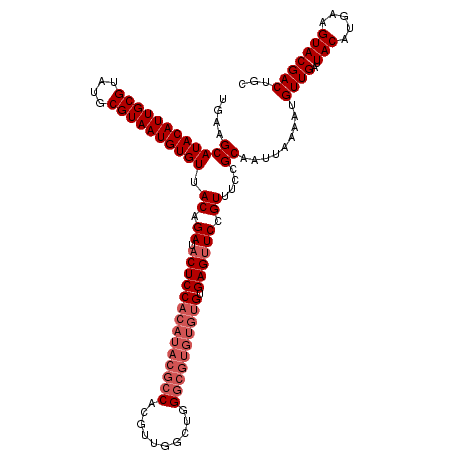
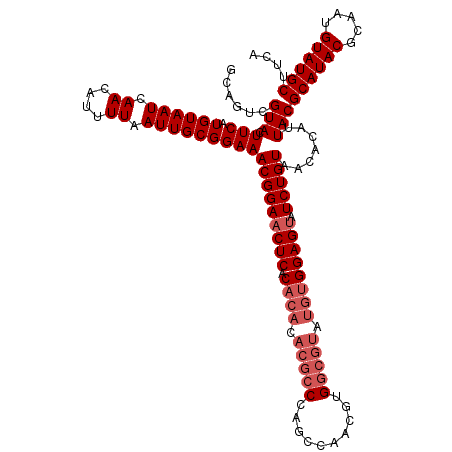
| Location | 17,123,254 – 17,123,374 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -26.99 |
| Energy contribution | -28.64 |
| Covariance contribution | 1.65 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123254 120 - 22407834 GCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAACGGAACUCGCACACACGCCCAGACAUCGUGGCGUAUGUGGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCAACA ......(((.(((.((((((.((....)).)))))))))(((((((((.((((.(((((..(....)..))))).)))))))).))))).......)))((((((.....)))))).... ( -34.50) >DroSec_CAF1 87075 100 - 1 GCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAACGGAACUCACACACACGC--------------------GGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCUUCA ((((..(((((((((((.....)))).....((.((....)).)).............--------------------)))))))))))..........((((((.....)))))).... ( -22.50) >DroSim_CAF1 103139 120 - 1 GCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAACGGAACUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCUUCA (((((........((((.....))))....))))).((((((((((((.((((.((((((.......).))))).)))))))).)))))..........((((((.....))))))))). ( -34.30) >DroEre_CAF1 89182 120 - 1 GCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAACGGAUCUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUUACACAUUACGCAUACGCAAUGUAUGCUACA ......(((......((((((..((.......))..))((((((((((.((((.((((((.......).))))).))))))).))))))).....))))((((((.....))))))))). ( -35.30) >DroYak_CAF1 88966 120 - 1 GCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAACGGAACUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUUACACAUUACGCAUACGCAAUGUAUGCUUCA (((((........((((.....))))....)))))(((((((((((((.((((.((((((.......).))))).)))))))).)))))).........((((((.....))))))))). ( -35.00) >consensus GCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAACGGAACUCACACACACGCCCAGCCAACGUGGCGUAUGUGGAGUAUCUGUAACACAUUACGCAUACGCAAUGUAUGCUUCA ......(((.(((.((((((.((....)).)))))))))(((((((((.((((.(((((..........))))).)))))))).))))).......)))((((((.....)))))).... (-26.99 = -28.64 + 1.65)

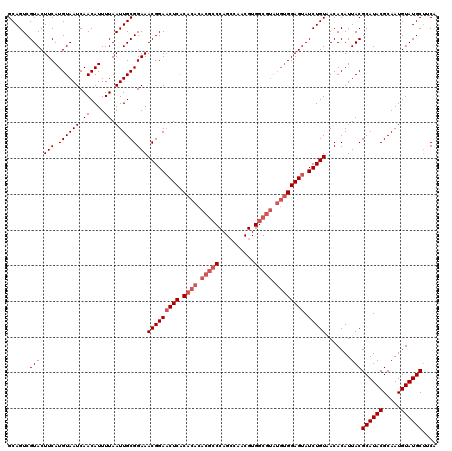
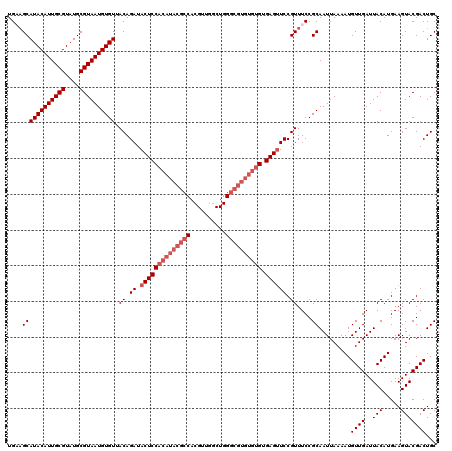
| Location | 17,123,334 – 17,123,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -33.66 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123334 120 - 22407834 AUAAUUUCCGUUUUCCUUUGUGGCUCCCACAGCAUCGGAUGGGUAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCGGCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAA ....(((((((((((((((((((((((((....(((.....)))....)))))))....))))))))))).......(((.....))).........................))))))) ( -33.40) >DroSec_CAF1 87135 120 - 1 AUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCGGCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAA ....(((((((((((((((((.((((((.((((.((....))))....)).)))))).))).))))))))).........(((((........((((.....))))....)))))))))) ( -36.50) >DroSim_CAF1 103219 120 - 1 AUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCGGCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAA ....(((((((((((((((((.((((((.((((.((....))))....)).)))))).))).))))))))).........(((((........((((.....))))....)))))))))) ( -36.50) >DroEre_CAF1 89262 120 - 1 AUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCGAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCGGCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAA ....(((((((((((((((((.((((((.((...(((......)))..)).)))))).))).))))))))).........(((((........((((.....))))....)))))))))) ( -40.10) >DroYak_CAF1 89046 120 - 1 AUAAUUUCCGUUUUCCUUUGUGGCACCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCGGCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAA ....((((((((((((((((((.(((((.((((.((....))))....))..)).))).))))))))))).......(((.....))).........................))))))) ( -31.60) >consensus AUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCGGCAGUCGUACUUCAUGUAAUCAACAUUUUAAUUGCGGAAA ....(((((((((((((((((.((((((.((((.((....))))....)).)))))).))).))))))))).........(((((........((((.....))))....)))))))))) (-33.66 = -33.90 + 0.24)

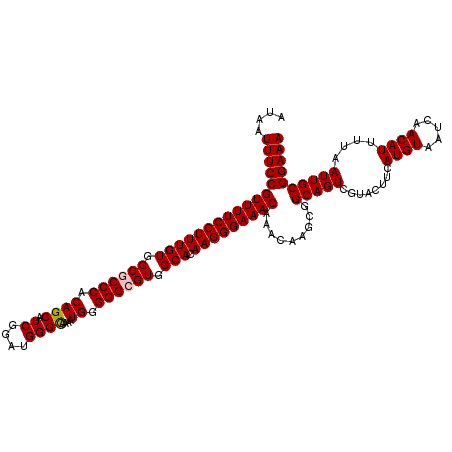
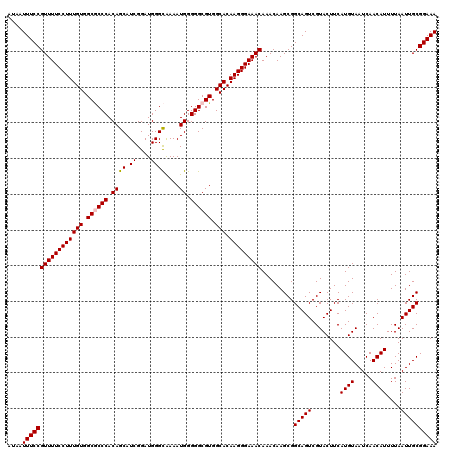
| Location | 17,123,374 – 17,123,494 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123374 120 - 22407834 AAUGAAAAGCUCAGCUAAGCGCCAACGCCCGCAUCUACACAUAAUUUCCGUUUUCCUUUGUGGCUCCCACAGCAUCGGAUGGGUAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCG .............(((..(.((((.(((((.(((.(((.((.....((((.......((((((...))))))...)))))).)))..))).))))))))).)...(....).....))). ( -36.20) >DroSec_CAF1 87175 120 - 1 AAUGAAAAGCUCAGCUAAGCGCCAACGCCCGCAUCUACACAUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCG .............(((..(.((((.(((((.(((...((((.................))))..((((((.......).)))))...))).))))))))).)...(....).....))). ( -37.13) >DroSim_CAF1 103259 120 - 1 AAUGAAAAGCUCAGCUAAGCGCCAACGCCCGCAUCUACACAUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCG .............(((..(.((((.(((((.(((...((((.................))))..((((((.......).)))))...))).))))))))).)...(....).....))). ( -37.13) >DroEre_CAF1 89302 120 - 1 AAUGAAAAGCUCAGCUAAGCGCCAACGCCCGCAUCUACACAUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCGAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCG ........(((..((...(((....)))..)).................((((((((((((.((((((.((...(((......)))..)).)))))).))).))))))))).....))). ( -39.00) >DroYak_CAF1 89086 120 - 1 AAUGAAAAGCUCAGCUAAGCGCCAACGCCCGCAUCUACACAUAAUUUCCGUUUUCCUUUGUGGCACCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCG .............(((..(.((((.((((((......).........((((((((((((((((...))))))........))).)))))))))))))))).)...(....).....))). ( -34.60) >consensus AAUGAAAAGCUCAGCUAAGCGCCAACGCCCGCAUCUACACAUAAUUUCCGUUUUCCUUUGUGGCGCCCACAGCAUCGGAUGGGCAAAAUGGGGGCGUGGCACAAGGGAAACAAACAAGCG .............(((..(.((((.((((((......).........((((((((((((((((...))))))........))).)))))))))))))))).)...(....).....))). (-34.66 = -34.50 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:50 2006