| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,122,937 – 17,123,177 |
| Length | 240 |
| Max. P | 0.999920 |

| Location | 17,122,937 – 17,123,057 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -39.00 |
| Energy contribution | -39.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17122937 120 + 22407834 UUCCCUCCGGGCGACCACCAUCAGCUAAGUAGAUUUGGGCUACCUGCAUGCAGCUAUGCUGCAGCGUUUAAGUGGGUAUAUAGUAUAUUCCUUUUUUUCCUUCGUGGUGCGGGAGGGAAA ((((((((((....)).(((....((....))...)))((.(((.((.((((((...))))))))....(((.(((((((...))))))))))............))))).)))))))). ( -40.40) >DroSec_CAF1 86757 118 + 1 UUCCCUCCGGGCGACCACCAUCAGCUAAGUAGAUUUGGGCUACCUGCAUGCAGCUAUGCUGCAGCGUUUAAGUGGGUAUAUAGUAUUUUCC-UUUUUUCCUUGGUGGUGCGGGAG-GAAA ((((..(((.((.(((((((....((....))...)))..((((..(.((((((...))))))........)..)))).............-.........)))).)).)))..)-))). ( -38.20) >DroSim_CAF1 102820 120 + 1 UUCCCUCCGGGCGACCACCAUCAGCUAAGUAGAUUUGGGCUACCUGCAUGCAGCUAUGCUGCAGCGUUUAAGUGGGUAUAUAGUAUAUUCCGUUUUUUCCUAGGUGGUGUGGGAGGGAAA ((((((((((....)).((((((.(((.(.(((..((((((((.(((.((((((...))))))))).....))))(((((...)))))))))..))).).))).))))).))))))))). ( -43.90) >DroEre_CAF1 88865 120 + 1 UUCCCUCCGGGCGACCACCAUCAGCUAAGUAGAUUUGGGCUACCUGCAUGCAGCUAUGCUGCAGCGUUUAAGUGGGUAUAUAGUAUAUUCCUUUUUUUGCUCGGCGGUGCGGGAGGGAAA ((((((((((....)).(((((.((..((((((...(((((((.(((.((((((...))))))))).....))))(((((...))))).)))...))))))..)))))).))))))))). ( -46.40) >DroYak_CAF1 88649 120 + 1 UUCCCUCCGGGCGACCACCAUCAGCUAAGUAGAUUUGGGCUACCUGCAUGCAGCUAUGCUGCAGCGUUUAAGUGGGUAUAUAGUAUAUUUCUUUUUUUACUCGGUGGUGCGGGAGGGAAA ((((((((..((.((((((.............((((((((...(((((.(((....)))))))).))))))))(((((...((.......)).....))))))))))))).)))))))). ( -45.50) >consensus UUCCCUCCGGGCGACCACCAUCAGCUAAGUAGAUUUGGGCUACCUGCAUGCAGCUAUGCUGCAGCGUUUAAGUGGGUAUAUAGUAUAUUCCUUUUUUUCCUCGGUGGUGCGGGAGGGAAA ((((((((..((.((((((...((((.((.....)).))))(((..(.((((((...))))))........)..))).........................)))))))).)))))))). (-39.00 = -39.44 + 0.44)

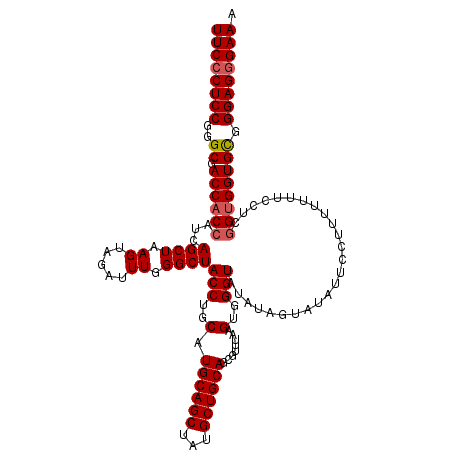

| Location | 17,122,937 – 17,123,057 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -37.14 |
| Energy contribution | -37.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17122937 120 - 22407834 UUUCCCUCCCGCACCACGAAGGAAAAAAAGGAAUAUACUAUAUACCCACUUAAACGCUGCAGCAUAGCUGCAUGCAGGUAGCCCAAAUCUACUUAGCUGAUGGUGGUCGCCCGGAGGGAA .((((((((.(((((((.(.((.......((.((((...)))).)).........((((((((...)))))).))((((((.......))))))..))..).))))).))..)))))))) ( -38.30) >DroSec_CAF1 86757 118 - 1 UUUC-CUCCCGCACCACCAAGGAAAAAA-GGAAAAUACUAUAUACCCACUUAAACGCUGCAGCAUAGCUGCAUGCAGGUAGCCCAAAUCUACUUAGCUGAUGGUGGUCGCCCGGAGGGAA .(((-((((.(((((((((.((......-..........................((((((((...)))))).))((((((.......))))))..))..))))))).))..))))))). ( -39.60) >DroSim_CAF1 102820 120 - 1 UUUCCCUCCCACACCACCUAGGAAAAAACGGAAUAUACUAUAUACCCACUUAAACGCUGCAGCAUAGCUGCAUGCAGGUAGCCCAAAUCUACUUAGCUGAUGGUGGUCGCCCGGAGGGAA .((((((((..(((((((..((.......((.((((...)))).)).........((((((((...)))))).))((((((.......))))))..))...)))))).)...)))))))) ( -38.80) >DroEre_CAF1 88865 120 - 1 UUUCCCUCCCGCACCGCCGAGCAAAAAAAGGAAUAUACUAUAUACCCACUUAAACGCUGCAGCAUAGCUGCAUGCAGGUAGCCCAAAUCUACUUAGCUGAUGGUGGUCGCCCGGAGGGAA .((((((((.((((((((((((.......((.((((...)))).)).........((((((((...)))))).))((((((.......)))))).)))..))))))).))..)))))))) ( -44.30) >DroYak_CAF1 88649 120 - 1 UUUCCCUCCCGCACCACCGAGUAAAAAAAGAAAUAUACUAUAUACCCACUUAAACGCUGCAGCAUAGCUGCAUGCAGGUAGCCCAAAUCUACUUAGCUGAUGGUGGUCGCCCGGAGGGAA .((((((((.((((((((((((.................................((((((((...)))))).))((((((.......)))))).)))..))))))).))..)))))))) ( -42.30) >consensus UUUCCCUCCCGCACCACCAAGGAAAAAAAGGAAUAUACUAUAUACCCACUUAAACGCUGCAGCAUAGCUGCAUGCAGGUAGCCCAAAUCUACUUAGCUGAUGGUGGUCGCCCGGAGGGAA .((((((((.(((((((((....................................((((((((...)))))).))((((((.......))))))......))))))).))..)))))))) (-37.14 = -37.62 + 0.48)

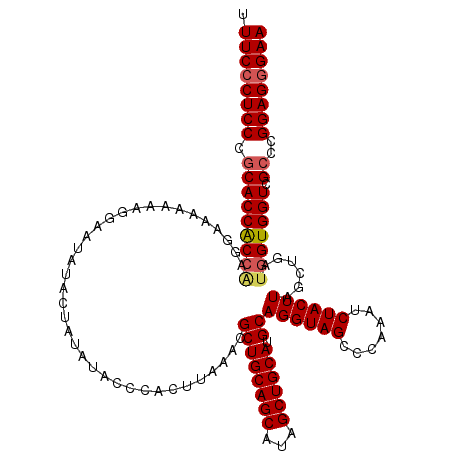

| Location | 17,123,057 – 17,123,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -22.00 |
| Energy contribution | -24.16 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17123057 120 - 22407834 UCUGGCAAUCGCAAAUCUAGCUGCCUCUCUUUUUCGCUGGAAUUCAUUACAUCAAAUAUAAAGAAAGUUUUAGGCAAUUUGCCAGAAGUCCUGGGAAAAGAGAAGAGCACCGAAAACCCA (((((((...((.......))))))((((((((((.(.(((.(((.............(((((....)))))(((.....))).))).))).)))))))))))))).............. ( -28.40) >DroSec_CAF1 86875 120 - 1 UCUGGCAAUCGCAAAUCUAGUUCCCUCUCUUUUUCGCAGGAAUUCAUUACAUCAAAUAUAAAGAAAGUUUUAGGCAAUUUGCCAGAAGUCCUGGGAAAAGAGAAGAGCAGCUAAAACCCA ....((....)).......((((..((((((((((.(((((.(((.............(((((....)))))(((.....))).))).))))))))))))))).))))............ ( -32.50) >DroSim_CAF1 102940 120 - 1 UCUGGCAAUCGCAAAUCUAGUUGCCUCUCUUUUGCGCUGGAAUUCAUUACAUCAAAUAUAAAGAAAGUUUUAGGCAAUUUGCCAGAAGUCCUGGGAAAAGAGAAGAGCAGCUAAAACCCA ....((....)).....((((((((((((((((.(.(.(((.(((.............(((((....)))))(((.....))).))).))).)).))))))).)).)))))))....... ( -31.40) >DroEre_CAF1 88985 120 - 1 CCUGGCAAUCGCAAAUCUAGCUGCGUCUCUUUCCCGCUGAAAUUCAUUACAUCAAAUAUAAAGAAAGUUUUAGGCAAUUUGCCAGAAGUCCCGGGAAAAGAGAAAAGCGACCAAAGCCCA ...(((....((.......))(((.(((((((((((..((..(((.............(((((....)))))(((.....))).))).)).)))).)))))))...)))......))).. ( -29.10) >DroYak_CAF1 88769 120 - 1 CCUGGCAAUCGCAAAUCUAGUUUCGUCUCUCUCUCGCUGAAAUUCAUUACAUCAAAUAUAAAGAAAGUUUUAGGCUAUUUGCCAGAAGUCCCGGGAAAAGAGAAAACCGACUAAAGCCCA ...(((....(.....)(((((...(((((.(((((..((..(((.............(((((....)))))(((.....))).))).)).)))))..))))).....)))))..))).. ( -20.10) >consensus UCUGGCAAUCGCAAAUCUAGUUGCCUCUCUUUUUCGCUGGAAUUCAUUACAUCAAAUAUAAAGAAAGUUUUAGGCAAUUUGCCAGAAGUCCUGGGAAAAGAGAAGAGCAACUAAAACCCA ....((....)).....(((((((.((((((((((.(.(((.(((.............(((((....)))))(((.....))).))).))).)))))))))))...)))))))....... (-22.00 = -24.16 + 2.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:45 2006