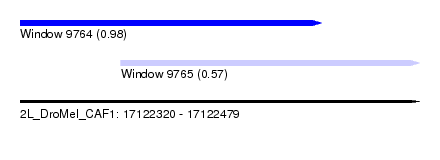
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,122,320 – 17,122,479 |
| Length | 159 |
| Max. P | 0.980379 |

| Location | 17,122,320 – 17,122,440 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -46.22 |
| Consensus MFE | -38.88 |
| Energy contribution | -40.48 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17122320 120 + 22407834 GCUUUCGUCUGCAUUGCUUAGCUACGAGCUGUGGCGCCUGGCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCUUCUGUAAGGCAAGCUGUGCCGAGAGUAAGGUGGCAAGCAGUU ........((((..((((..(((((.....)))))....))))((((((((......((((((((....((((((((((....)))))))))).))))))))....)))))))))))).. ( -51.10) >DroSec_CAF1 86127 120 + 1 GCUUUCGUCUGCAUUGCUUAGCUACGAGCUGUGGCGCCUGGCAUUGUCGCUCAAGUGUUUCGGCAAACUGACUUGCCAUCUGUAAGGCAAGCUGUGCUGAAAGUAAGGUGGCAAGCAGUU ....((((..((........)).))))((((..((((.((((......)).)).))))..)))).(((((.((((((((((....((((.....)))).......))))))))))))))) ( -44.40) >DroSim_CAF1 102192 120 + 1 GCUUUCGUCUGCAUUGCUUAGCUACGAGCUGUGGCGCCUGGCAUUGUCGCUCAAGUGUUUCGGCAAACUGACUUGCCUUCUGUAAGGCAAGCUGUGCUGAAAGUAAGGUGGCAAGCAGUU ........((((..((((..(((((.....)))))....))))((((((((......((((((((....(.((((((((....)))))))))..))))))))....)))))))))))).. ( -44.60) >DroEre_CAF1 88260 120 + 1 GCUUUCGUCUGCAUUGCUUGGCUCCGAGCUGUGGCGCCUCGCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCGUCUGUGCGGCAAGCUGUGCAGAAAGCAAGGUGGCAAGAAGUU ((...((((.(((..(((((....))))))))))))....)).((((((((....((((((.(((....((((((((((....)))))))))).))).).))))).))))))))...... ( -46.00) >DroYak_CAF1 88021 120 + 1 GCUUUCGUCUGCAUUGCUUGGCUCCGUGCAGUGGGGUCUGGCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCAUCUGUAAGGCAAGCUGUGCAGAAAGCAAGGUGGCAAGAAGUU (((((.........((((.(((((((.....))))))).))))((((((((....((((((.(((....((((((((........)))))))).))).).))))).))))))))))))). ( -45.00) >consensus GCUUUCGUCUGCAUUGCUUAGCUACGAGCUGUGGCGCCUGGCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCAUCUGUAAGGCAAGCUGUGCAGAAAGUAAGGUGGCAAGCAGUU ........((((..((((..(((((.....)))))....))))((((((((....((((((((((....((((((((((....)))))))))).)))))))).)).)))))))))))).. (-38.88 = -40.48 + 1.60)


| Location | 17,122,360 – 17,122,479 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.81 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.46 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17122360 119 + 22407834 GCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCUUCUGUAAGGCAAGCUGUGCCGAGAGUAAGGUGGCAAGCAGUUUCAGGAC-UGAAGGGAGCGGUCUGAAGGAUAUCUGUCUCU ...((((((((......((((((((....((((((((((....)))))))))).))))))))....))))))))((((..((.((((-((.......))))))....))...)))).... ( -46.50) >DroSec_CAF1 86167 119 + 1 GCAUUGUCGCUCAAGUGUUUCGGCAAACUGACUUGCCAUCUGUAAGGCAAGCUGUGCUGAAAGUAAGGUGGCAAGCAGUUGCAGGAC-UGAAGGGAGCGGUCUGAAGGAUAUCUGUCUCU ......((((((....((((..((((.(((.((((((((((....((((.....)))).......))))))))))))))))).))))-......))))))......((((....)))).. ( -39.20) >DroSim_CAF1 102232 119 + 1 GCAUUGUCGCUCAAGUGUUUCGGCAAACUGACUUGCCUUCUGUAAGGCAAGCUGUGCUGAAAGUAAGGUGGCAAGCAGUUGGAGGAC-UGAAGGGAGCGGUCUGAAGGAUAUCUGUCUCU ((.((((((((......((((((((....(.((((((((....)))))))))..))))))))....))))))))))....(((((((-((.......)))))....((....))..)))) ( -38.50) >DroEre_CAF1 88300 120 + 1 GCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCGUCUGUGCGGCAAGCUGUGCAGAAAGCAAGGUGGCAAGAAGUUGCAGGAUGUGGACGAAGCGGUCUAAAGGAUAUCUGGCCUU ...((((((((....((((((.(((....((((((((((....)))))))))).))).).))))).))))))))......((.((((((((((......)))).....)))))).))... ( -41.80) >DroYak_CAF1 88061 120 + 1 GCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCAUCUGUAAGGCAAGCUGUGCAGAAAGCAAGGUGGCAAGAAGUUGCAGGACGUAGAAGGAGCGGCCUGAAGGAUAUCUGCCCCU ((.....(((....)))((((.(((....((((((((........)))))))).))).)))))).(((.((((.((.(((.((((.(((.......))).))))...))).))))))))) ( -42.00) >consensus GCAUUGUCGCUCAAGUGUUUCGGCAAACUGGCUUGCCAUCUGUAAGGCAAGCUGUGCAGAAAGUAAGGUGGCAAGCAGUUGCAGGAC_UGAAGGGAGCGGUCUGAAGGAUAUCUGUCUCU ((.((((((((....((((((((((....((((((((((....)))))))))).)))))))).)).)))))))))).................(((((((............))).)))) (-31.58 = -32.46 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:42 2006