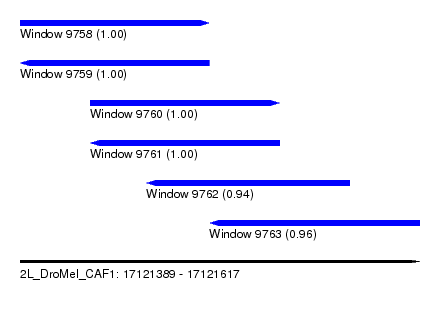
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,121,389 – 17,121,617 |
| Length | 228 |
| Max. P | 0.999346 |

| Location | 17,121,389 – 17,121,497 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -33.80 |
| Energy contribution | -33.48 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -5.05 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17121389 108 + 22407834 ACAUUACACUCGACCGAUUUCCAGGAGAGUCAAAAAGGUAAUUUCUAGAGAAACGGUCACGGAAAAAGAAUU------------UCCGUGACGUUUCUUUGGAGCUUACCAAAGAUUUCU .......((((..((........)).))))......(((((.(((((((((((((.(((((((((.....))------------)))))))))))))))))))).))))).......... ( -41.80) >DroSec_CAF1 85245 108 + 1 ACAUGACACUAGACCGAUUUCCAGGAGAGUCCAAAGGGUAAUUUCUAGAGAAACGAUCACGGAGAGUGAAUU------------UCCGUGACGUUUCUUUAGAGCUUACCAAAGAUUUCU ...............((..((..((.....))....(((((.(((((((((((((.(((((((((.....))------------)))))))))))))))))))).)))))...))..)). ( -41.30) >DroSim_CAF1 101309 108 + 1 ACAUCACACUAGACCGAUUUCCAGGAGAGUCCAAAAGGUAAUUUCUAGAGAAACGAUCACGGAGAGUAAAUU------------UCCGUGACGUUUCUUUAGAGCUUACCAAAGAUUUUU ..(((..(((...((........))..)))......(((((.(((((((((((((.(((((((((.....))------------)))))))))))))))))))).)))))...))).... ( -40.60) >DroEre_CAF1 87346 120 + 1 ACAUUGCACUAGACCGAUUUGCAGGAGAGUCGAAAAGGUAGUUUCUAGAGAAACCAUCACGGAAAGAAAAUUUUUGACUUAAUCUCCGUGACGUUUCUUUAGAGCUUGCCAAAGUGUUCU .....(((((....((((((......))))))....(((((.((((((((((((..(((((((.....................))))))).)))))))))))).)))))..)))))... ( -38.90) >DroYak_CAF1 87050 120 + 1 ACAUGACACUAGACCGAUUUGCAGGAGAAUCGAAAAGGUAAUUUCUAGAGAAACGAUCAAGGAAAUACAAUUUUUGACUUGAUCUCCGUGACGUUUCUUUAGAGCUUGCCUGAGAUUUCU .............((........)).(((((....((((((.(((((((((((((.(((.(((.((.(((........))))).))).)))))))))))))))).))))))..))))).. ( -36.10) >consensus ACAUGACACUAGACCGAUUUCCAGGAGAGUCCAAAAGGUAAUUUCUAGAGAAACGAUCACGGAAAGAAAAUU____________UCCGUGACGUUUCUUUAGAGCUUACCAAAGAUUUCU ...............(((((......))))).....(((((.(((((((((((((.(((((((.....................)))))))))))))))))))).))))).......... (-33.80 = -33.48 + -0.32)

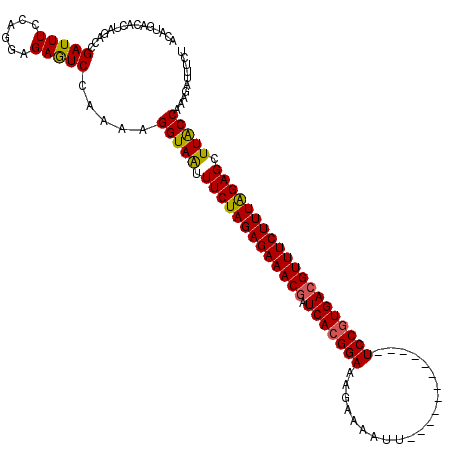
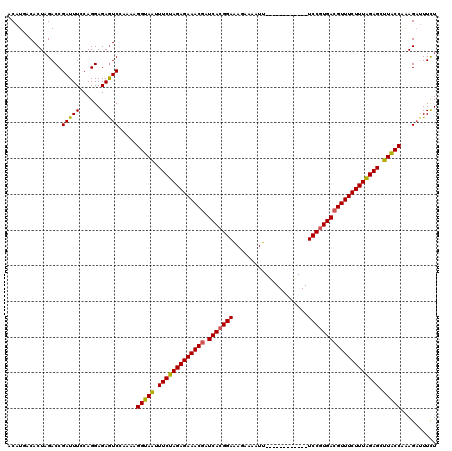
| Location | 17,121,389 – 17,121,497 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -24.80 |
| Energy contribution | -26.08 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17121389 108 - 22407834 AGAAAUCUUUGGUAAGCUCCAAAGAAACGUCACGGA------------AAUUCUUUUUCCGUGACCGUUUCUCUAGAAAUUACCUUUUUGACUCUCCUGGAAAUCGGUCGAGUGUAAUGU ..........(((((..((...((((((((((((((------------((.....))))))))).)))))))...))..)))))......((((..(((.....)))..))))....... ( -34.80) >DroSec_CAF1 85245 108 - 1 AGAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA------------AAUUCACUCUCCGUGAUCGUUUCUCUAGAAAUUACCCUUUGGACUCUCCUGGAAAUCGGUCUAGUGUCAUGU ..........(((((..((((.((((((((((((((------------.........))))))).))))))).))))..))))).....(((....(((((......))))).))).... ( -35.60) >DroSim_CAF1 101309 108 - 1 AAAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA------------AAUUUACUCUCCGUGAUCGUUUCUCUAGAAAUUACCUUUUGGACUCUCCUGGAAAUCGGUCUAGUGUGAUGU .....(((..(((((..((((.((((((((((((((------------.........))))))).))))))).))))..)))))....))).((.((((((......))))).).))... ( -36.00) >DroEre_CAF1 87346 120 - 1 AGAACACUUUGGCAAGCUCUAAAGAAACGUCACGGAGAUUAAGUCAAAAAUUUUCUUUCCGUGAUGGUUUCUCUAGAAACUACCUUUUCGACUCUCCUGCAAAUCGGUCUAGUGCAAUGU ....((((..(((....((((.((((((((((((((((..((((.....))))..)))))))))).)))))).))))...........(((............)))))).))))...... ( -30.60) >DroYak_CAF1 87050 120 - 1 AGAAAUCUCAGGCAAGCUCUAAAGAAACGUCACGGAGAUCAAGUCAAAAAUUGUAUUUCCUUGAUCGUUUCUCUAGAAAUUACCUUUUCGAUUCUCCUGCAAAUCGGUCUAGUGUCAUGU .((...((.((((....((((.((((((((((.(((((((((........))).)))))).))).))))))).))))...........(((((........)))))))))))..)).... ( -31.70) >consensus AGAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA____________AAUUCUCUUUCCGUGAUCGUUUCUCUAGAAAUUACCUUUUCGACUCUCCUGGAAAUCGGUCUAGUGUAAUGU ..........(((((..((((.((((((((((((((.....................))))))).))))))).))))..)))))..(((((((............)))))))........ (-24.80 = -26.08 + 1.28)

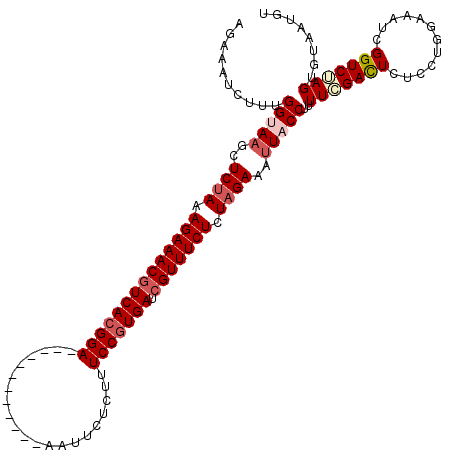
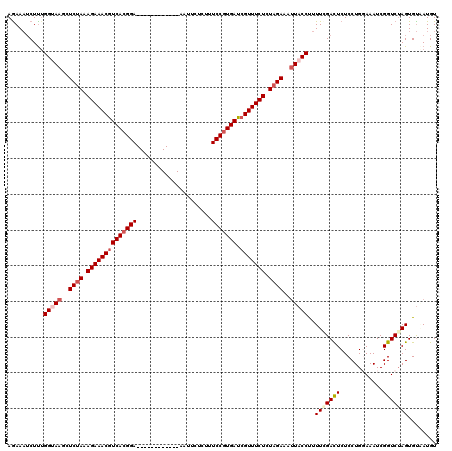
| Location | 17,121,429 – 17,121,537 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -27.76 |
| Energy contribution | -29.16 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
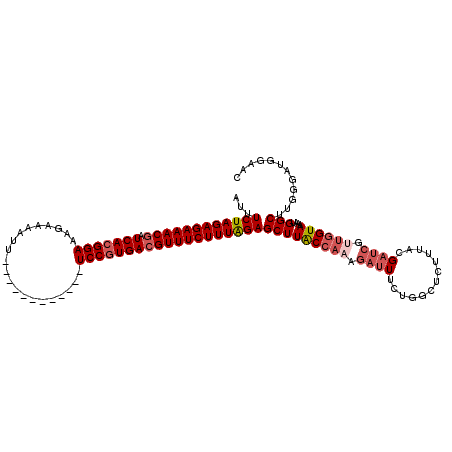
>2L_DroMel_CAF1 17121429 108 + 22407834 AUUUCUAGAGAAACGGUCACGGAAAAAGAAUU------------UCCGUGACGUUUCUUUGGAGCUUACCAAAGAUUUCUGGCUCUUUGCGAUCGUUGGUAAAAUGGCCUGGGAUGGAAC ..(((((((((((((.(((((((((.....))------------))))))))))))))))))))....(((....(..(.((((.(((((.(....).)))))..)))).)..))))... ( -41.20) >DroSec_CAF1 85285 108 + 1 AUUUCUAGAGAAACGAUCACGGAGAGUGAAUU------------UCCGUGACGUUUCUUUAGAGCUUACCAAAGAUUUCUGGCUCUUUACGAUCGUUGGUAAAAUGGCUUGGGAUGGAAC ..(((((((((((((.(((((((((.....))------------))))))))))))))))))))....(((....(..(.((((.(((((.(....).)))))..)))).)..))))... ( -40.50) >DroSim_CAF1 101349 108 + 1 AUUUCUAGAGAAACGAUCACGGAGAGUAAAUU------------UCCGUGACGUUUCUUUAGAGCUUACCAAAGAUUUUUGGCUCUUUACGAUCGUUGGUAAAAUGGCUUGGGAUGGAAC ..(((((((((((((.(((((((((.....))------------))))))))))))))))))))....(((....(((..((((.(((((.(....).)))))..))))..))))))... ( -39.00) >DroEre_CAF1 87386 120 + 1 GUUUCUAGAGAAACCAUCACGGAAAGAAAAUUUUUGACUUAAUCUCCGUGACGUUUCUUUAGAGCUUGCCAAAGUGUUCUCUUUCCUUUCGAUCGGUCGUAGAAUGGCUUGGGAUGAAAC ..((((((((((((..(((((((.....................))))))).))))))))))))....((((..((((((....((........))....))))))..))))........ ( -31.20) >DroYak_CAF1 87090 120 + 1 AUUUCUAGAGAAACGAUCAAGGAAAUACAAUUUUUGACUUGAUCUCCGUGACGUUUCUUUAGAGCUUGCCUGAGAUUUCUCUCUCUUUUUGAUAGAUGGUAAAAUGGCAUGGGAUGGAAC ...((((((((((((.(((.(((.((.(((........))))).))).)))))))))))))))(((((((.((((....))))(((.......))).))))....)))............ ( -30.90) >consensus AUUUCUAGAGAAACGAUCACGGAAAGAAAAUU____________UCCGUGACGUUUCUUUAGAGCUUACCAAAGAUUUCUGGCUCUUUACGAUCGUUGGUAAAAUGGCUUGGGAUGGAAC ...((((((((((((.(((((((.....................)))))))))))))))))))(((((((((.((((.............)))).))))))....)))............ (-27.76 = -29.16 + 1.40)

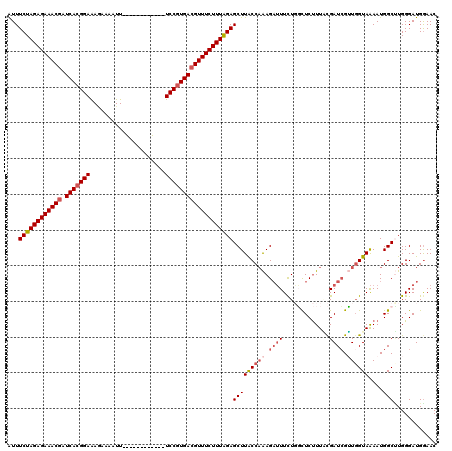
| Location | 17,121,429 – 17,121,537 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.87 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17121429 108 - 22407834 GUUCCAUCCCAGGCCAUUUUACCAACGAUCGCAAAGAGCCAGAAAUCUUUGGUAAGCUCCAAAGAAACGUCACGGA------------AAUUCUUUUUCCGUGACCGUUUCUCUAGAAAU .(((.......((....((((((((.(((..(.........)..))).))))))))..))..((((((((((((((------------((.....))))))))).)))))))...))).. ( -31.50) >DroSec_CAF1 85285 108 - 1 GUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCCAGAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA------------AAUUCACUCUCCGUGAUCGUUUCUCUAGAAAU ............((..((((((........)))))).((((((....))))))..))((((.((((((((((((((------------.........))))))).))))))).))))... ( -30.90) >DroSim_CAF1 101349 108 - 1 GUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCCAAAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA------------AAUUUACUCUCCGUGAUCGUUUCUCUAGAAAU ............((..((((((........)))))).((((((....))))))..))((((.((((((((((((((------------.........))))))).))))))).))))... ( -30.80) >DroEre_CAF1 87386 120 - 1 GUUUCAUCCCAAGCCAUUCUACGACCGAUCGAAAGGAAAGAGAACACUUUGGCAAGCUCUAAAGAAACGUCACGGAGAUUAAGUCAAAAAUUUUCUUUCCGUGAUGGUUUCUCUAGAAAC ............((((((((....((........))....)))).....))))....((((.((((((((((((((((..((((.....))))..)))))))))).)))))).))))... ( -33.70) >DroYak_CAF1 87090 120 - 1 GUUCCAUCCCAUGCCAUUUUACCAUCUAUCAAAAAGAGAGAGAAAUCUCAGGCAAGCUCUAAAGAAACGUCACGGAGAUCAAGUCAAAAAUUGUAUUUCCUUGAUCGUUUCUCUAGAAAU ...........((((.........(((.......)))((((....)))).))))...((((.((((((((((.(((((((((........))).)))))).))).))))))).))))... ( -35.70) >consensus GUUCCAUCCCAAGCCAUUUUACCAACGAUCGAAAAGAGCCAGAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA____________AAUUCUCUUUCCGUGAUCGUUUCUCUAGAAAU ............((((............((.....))............))))....((((.((((((((((((((.....................))))))).))))))).))))... (-22.31 = -22.87 + 0.56)

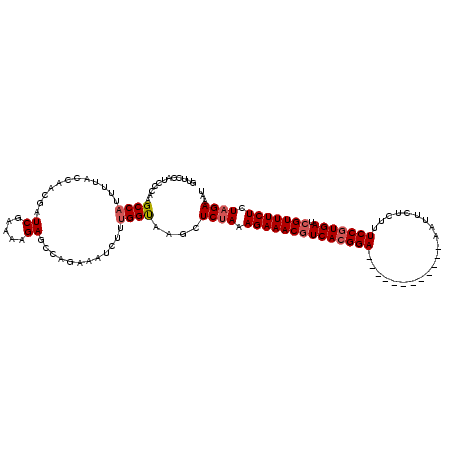
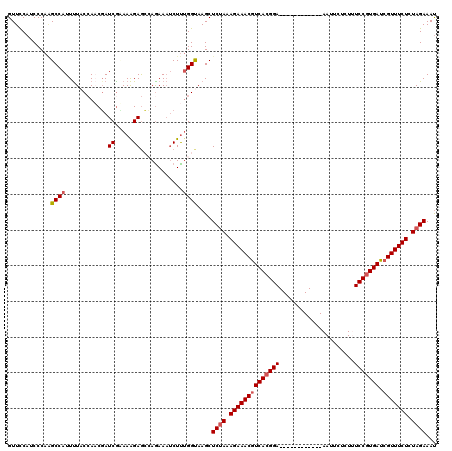
| Location | 17,121,461 – 17,121,577 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.77 |
| Mean single sequence MFE | -28.49 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17121461 116 - 22407834 AAAUAAACCACGUAAAGUGGCUAAAAAGCCUCAACAGGCGGUUCCAUCCCAGGCCAUUUUACCAACGAUCGCAAAGAGCCAGAAAUCUUUGGUAAGCUCCAAAGAAACGUCACGGA---- .......((..(((((((((((.....((((....))))((......))..)))))))))))..(((...((.....))......(((((((......)))))))..)))...)).---- ( -33.50) >DroSec_CAF1 85317 116 - 1 AAAUAAACCACGUAAAGUGGUUAAAAAGCCUCAAAAGACGGUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCCAGAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA---- .....((((((.....))))))..............((((((((.............(((((........)))))))))).....(((((((......)))))))...))).....---- ( -23.51) >DroSim_CAF1 101381 116 - 1 AAAUAAACCACGUAAAGUGGUUAAAAAGCCUCAAAAGGCGGUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCCAAAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA---- .......((..(((((((((((.....((((....))))((......))..)))))))))))....((.(((.....((((((....))))))....((....)).)))))..)).---- ( -28.20) >DroEre_CAF1 87426 120 - 1 AAAUAAGCCACGUAAAGUGGCCAAAAAGCCACAAAAGGAGGUUUCAUCCCAAGCCAUUCUACGACCGAUCGAAAGGAAAGAGAACACUUUGGCAAGCUCUAAAGAAACGUCACGGAGAUU ......((((((((..(((((......)))))....((((((((......))))).))))))).....((....)).............))))...((((...((....))..))))... ( -26.30) >DroYak_CAF1 87130 120 - 1 AAAUAAGUCACGUAAAGUGGCCACAAAGCCCCGAGAGGCGGUUCCAUCCCAUGCCAUUUUACCAUCUAUCAAAAAGAGAGAGAAAUCUCAGGCAAGCUCUAAAGAAACGUCACGGAGAUC ......(((..((((((((((.....((((((....)).)))).........))))))))))...............((((....)))).)))...((((...((....))..))))... ( -30.94) >consensus AAAUAAACCACGUAAAGUGGCUAAAAAGCCUCAAAAGGCGGUUCCAUCCCAAGCCAUUUUACCAACGAUCGAAAAGAGCCAGAAAUCUUUGGUAAGCUCUAAAGAAACGUCACGGA____ .......((..((((((((((......(((......)))((......))...)))))))))).......................(((((((......)))))))........))..... (-21.24 = -21.88 + 0.64)

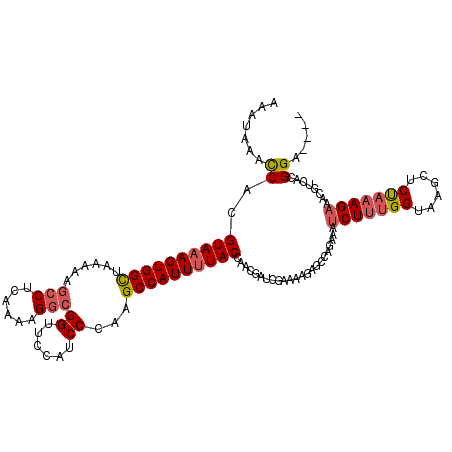
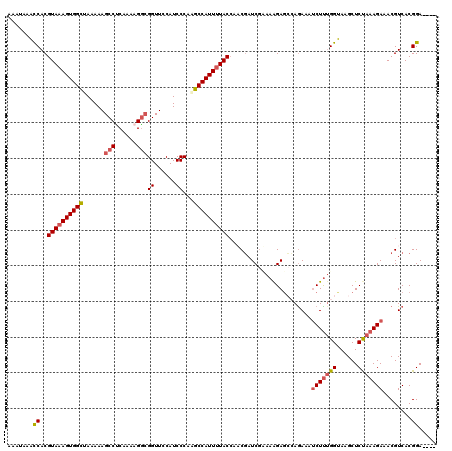
| Location | 17,121,497 – 17,121,617 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.97 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -20.30 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17121497 120 - 22407834 AAAAAUCAAAACCUCAAAGGAUUACAAAGACAACGGUUGAAAAUAAACCACGUAAAGUGGCUAAAAAGCCUCAACAGGCGGUUCCAUCCCAGGCCAUUUUACCAACGAUCGCAAAGAGCC ............(((...(((((...........((((.......))))..(((((((((((.....((((....))))((......))..)))))))))))....)))).)...))).. ( -28.20) >DroSec_CAF1 85353 120 - 1 AAAGAUCAAAAGCUCAAAGGAUUACAAAGACAGUGGUAGAAAAUAAACCACGUAAAGUGGUUAAAAAGCCUCAAAAGACGGUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCC ...........((((..(.((((.........(((((.........)))))(((((((((((....((((((....)).))))........)))))))))))....)))).)...)))). ( -27.20) >DroSim_CAF1 101417 120 - 1 AAAGAUCAAAACCUCAAAAGAUUACAAAGACACCGGUUGAAAAUAAACCACGUAAAGUGGUUAAAAAGCCUCAAAAGGCGGUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCC ...((((...........................((((.......))))..(((((((((((.....((((....))))((......))..)))))))))))....)))).......... ( -27.70) >DroYak_CAF1 87170 120 - 1 AAAGACCAAAGGCGCAAGGCGUUACAAAGACUAUGGUUGGAAAUAAGUCACGUAAAGUGGCCACAAAGCCCCGAGAGGCGGUUCCAUCCCAUGCCAUUUUACCAUCUAUCAAAAAGAGAG ...(((((..(.(....).)(((.....)))..))))).............((((((((((.....((((((....)).)))).........)))))))))).................. ( -28.34) >consensus AAAGAUCAAAACCUCAAAGGAUUACAAAGACAACGGUUGAAAAUAAACCACGUAAAGUGGCUAAAAAGCCUCAAAAGGCGGUUCCAUCCCAAGCCAUUUUACCAACGAUCGUAAAGAGCC ...((((...........................((((.......))))..(((((((((((.....(((......)))((......))..)))))))))))....)))).......... (-20.30 = -21.05 + 0.75)

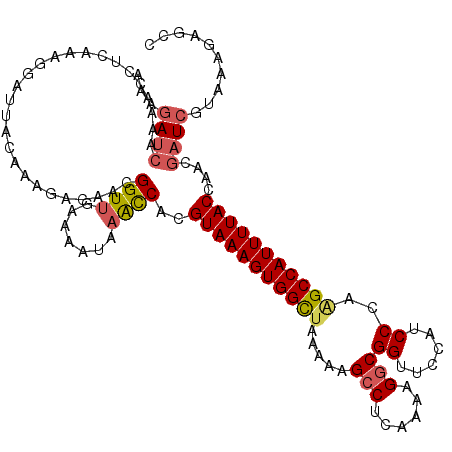
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:39 2006