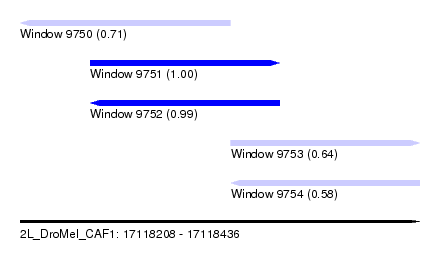
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,118,208 – 17,118,436 |
| Length | 228 |
| Max. P | 0.997650 |

| Location | 17,118,208 – 17,118,328 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.06 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
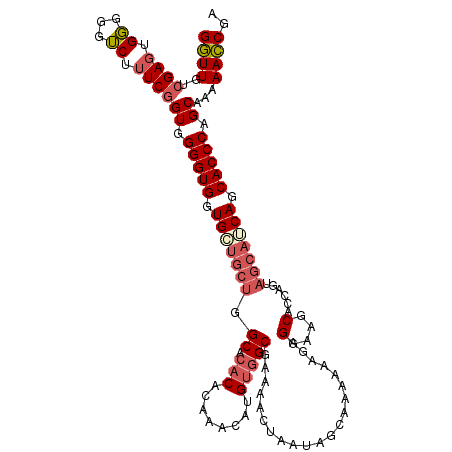
>2L_DroMel_CAF1 17118208 120 - 22407834 ACCAAAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCCGAACUAAGCAGCUCUGGCCCAUGGCUCCCCAUCUCCCACC .............((((((((((...(((((((((((((...(.((((...)))).)......))))).)))))).))...))))...))))))(((...((((....))))...))).. ( -35.60) >DroSec_CAF1 82053 120 - 1 ACCAAAACCAACCGAGUCGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCCGAACUAAGCAGCCCUGGCCCAUGGCUCCCCAUCUCCCACC .............(((((((((((..(((...(((((((...(.((((...)))).)......))))))).((((....)))).......))).))))))..)))))............. ( -34.00) >DroSim_CAF1 98118 118 - 1 ACCAAAACCAACUGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGA-UUUGCAGCACCCUUUGACCCCGAAAAAAACAAACCUGGCCCAUGGUUC-CCAUCUCCCAAC ...............((((((...(((((((((((((((...(.((((...)))).).-....))))).))))))...............((((........)))).-))))..)))))) ( -34.10) >DroEre_CAF1 84098 119 - 1 ACCAAAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGGUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCCGAACUAAGCAGCCCUUGCCCAUGGCUCC-CAUCUUCCACC .............(((((((((....(((...(((((((..(((((....)))))........))))))).((((....)))).......)))...)))))..)))).-........... ( -35.70) >DroYak_CAF1 83971 119 - 1 ACCAGAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGGUGCUCCACCUGAAUUUGCAGCACCCUUUGACCCCGAACUAAGCAGCCCUUGCCCAUGACUCC-CCAUCCCCACC .............(((((((((....(((...(((((((...(.((((...)))).)......))))))).((((....)))).......)))...)))))..)))).-........... ( -32.10) >consensus ACCAAAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCCGAACUAAGCAGCCCUGGCCCAUGGCUCCCCAUCUCCCACC .............(((((((((((..(((...(((((((..((((......))))........))))))).((((....)))).......))).))))))..)))))............. (-27.70 = -28.06 + 0.36)

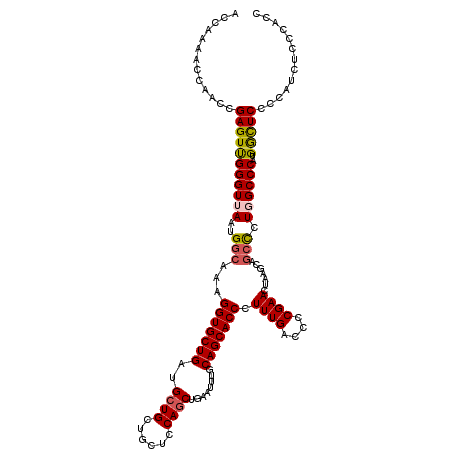
| Location | 17,118,248 – 17,118,356 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -47.84 |
| Consensus MFE | -38.70 |
| Energy contribution | -39.62 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17118248 108 + 22407834 GGGGUCAAAGGGUGCUGCAAAUUCAGCUGGAGCAGCAGCAUCAGCACCUUUGCCAUUAACCCAACUCGGUUGGUUUUGGUUCGGUUUUUGCUGGGUGCUG------------GUGCUUCC ((((........((((((...(((....)))))))))((((((((((((..((.(..((((.(((.(((......)))))).))))..))).))))))))------------)))))))) ( -41.30) >DroSec_CAF1 82093 117 + 1 GGGGUCAAAGGGUGCUGCAAAUUCAGCUGGAGCAGCAGCAUCAGCACCUUUGCCAUUAACCCGACUCGGUUGGUUUUGGUUCGGUUUUUGCUGGGUGCUGAUGCUAC---UGGUGCUUCC ((((.((..((..(((((...(((....))))))))(((((((((((((..((.(..((((.(((.(((......)))))).))))..))).))))))))))))).)---)..)))))). ( -47.10) >DroSim_CAF1 98157 116 + 1 GGGGUCAAAGGGUGCUGCAAA-UCAGCUGGAGCAGCAGCAUCAGCACCUUUGCCAUUAACCCAACUCAGUUGGUUUUGGUUUGGUUUUUGCUGGGUGCUGCUGCUUC---GAGUGCUUCC (((((........((((....-.))))(((((((((((((((((((((...((((.....(((((...)))))...))))..)))....))).))))))))))))))---)...))))). ( -47.60) >DroEre_CAF1 84137 117 + 1 GGGGUCAAAGGGUGCUGCAAAUUCAGCUGGAGCACCAGCAUCAGCACCUUUGCCAUUAACCCAACUCGGUUGGUUUUGGUUCGCUUUUUGCUGGGUGCUGAUGCUAC---UGGUGCUUCC (((((...((((((((.((........)).))))))(((((((((((((..((((.....(((((...)))))...))))..((.....)).))))))))))))).)---)...))))). ( -49.80) >DroYak_CAF1 84010 120 + 1 GGGGUCAAAGGGUGCUGCAAAUUCAGGUGGAGCACCAGCAUCAGCACCUUUGCCAUUAACCCAACUCGGUUGGUUCUGGUUCGGUUUUUGCUGGGUGCUGAUGCUACCUCUGGUGCUUCC (((((.....((((((.((........)).))))))(((((((((((((..((.(..((((.(((.(((......)))))).))))..))).)))))))))))))))))).......... ( -53.40) >consensus GGGGUCAAAGGGUGCUGCAAAUUCAGCUGGAGCAGCAGCAUCAGCACCUUUGCCAUUAACCCAACUCGGUUGGUUUUGGUUCGGUUUUUGCUGGGUGCUGAUGCUAC___UGGUGCUUCC (((((.....(.((((.((........)).)))).)(((((((((((((..((.(..((((.(((.(((......)))))).))))..))).))))))))))))).........))))). (-38.70 = -39.62 + 0.92)

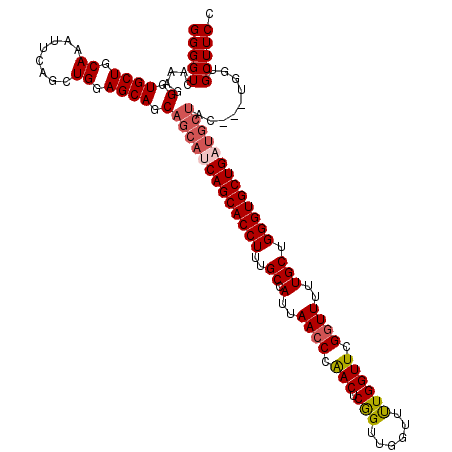
| Location | 17,118,248 – 17,118,356 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -42.76 |
| Consensus MFE | -32.24 |
| Energy contribution | -34.12 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17118248 108 - 22407834 GGAAGCAC------------CAGCACCCAGCAAAAACCGAACCAAAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCC (((.(((.------------(((((..((((....(((...(((...(((((...))))).....)))....))))))).))))).)))))).((((......))))............. ( -35.40) >DroSec_CAF1 82093 117 - 1 GGAAGCACCA---GUAGCAUCAGCACCCAGCAAAAACCGAACCAAAACCAACCGAGUCGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCC (((.(((...---((((((((((((((..((.....((((..(..........)..)))).......))...)))))))))))))))))))).((((......))))............. ( -44.80) >DroSim_CAF1 98157 116 - 1 GGAAGCACUC---GAAGCAGCAGCACCCAGCAAAAACCAAACCAAAACCAACUGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGA-UUUGCAGCACCCUUUGACCCC ((........---(.((((((((((..((((....(((...(((...(((((...))))).....)))....))))))).)))))))))).).((((.-....))))..))......... ( -39.30) >DroEre_CAF1 84137 117 - 1 GGAAGCACCA---GUAGCAUCAGCACCCAGCAAAAAGCGAACCAAAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGGUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCC (((.((((((---...(((((((((((..((.....))...(((...(((((...))))).....)))....)))))))))))))))))))).((((......))))............. ( -48.50) >DroYak_CAF1 84010 120 - 1 GGAAGCACCAGAGGUAGCAUCAGCACCCAGCAAAAACCGAACCAGAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGGUGCUCCACCUGAAUUUGCAGCACCCUUUGACCCC ........(((((((((((((((((((..((...((((.(((..(.......)..))).))))....))...)))))))))))))(((((.((........)).)))))))))))..... ( -45.80) >consensus GGAAGCACCA___GUAGCAUCAGCACCCAGCAAAAACCGAACCAAAACCAACCGAGUUGGGUUAAUGGCAAAGGUGCUGAUGCUGCUGCUCCAGCUGAAUUUGCAGCACCCUUUGACCCC (((.(((.(......((((((((((((..((...((((.((((..........).))).))))....))...))))))))))))).)))))).((((......))))............. (-32.24 = -34.12 + 1.88)


| Location | 17,118,328 – 17,118,436 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -22.60 |
| Energy contribution | -24.85 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17118328 108 + 22407834 UCGGUUUUUGCUGGGUGCUG---------GUGCUUCCUCUUUUUUGCUAUUAGUUUUCGCCACAUGUUUGUGUGCGCCUGCAGCACCACCCCACCGAAAGACCCCCACUCGACAACC ..((((((((.((((.(.((---------(((((....(......)......((...(((((((....)))).)))...)))))))))))))).).))))))).............. ( -31.30) >DroSec_CAF1 82173 117 + 1 UCGGUUUUUGCUGGGUGCUGAUGCUACUGGUGCUUCCUCUUUUUUGCUAUUAGUUUUCGCCACAUGUUUGUGUGUGCCAGCAUCACCACCCCACCGAAAGACCCCCACUCGACAACC ..((((((((..(((((.(((((((((((((((............)).))))))....(((((((....))))).)).))))))).)))))...).))))))).............. ( -36.10) >DroSim_CAF1 98236 117 + 1 UUGGUUUUUGCUGGGUGCUGCUGCUUCGAGUGCUUCCUCUUUUUUGUUAUUAGUUUUCGCGACAUGUUUGUGUGUGCCAGCAGCACCACCCCACCGAAAGACCCCCACUCGACAACC ..((((((((..(((((.(((((((..(((.(((....(......).....))).)))(((((((....)))).))).))))))).)))))...).))))))).............. ( -32.80) >DroEre_CAF1 84217 115 + 1 UCGCUUUUUGCUGGGUGCUGAUGCUACUGGUGCUUCCUCUUUUUUGCUAUUAGUUUUCGCCGCUUGUUUGUGUGUGCCAGCAGCAACACCCCACCGAAAGCCCCAC--UCUACAAGC ..((((((.(.((((.(.((.((((.((((..(............((...........))(((......))).)..)))).)))).)))))))).)))))).....--......... ( -30.50) >consensus UCGGUUUUUGCUGGGUGCUGAUGCUACUGGUGCUUCCUCUUUUUUGCUAUUAGUUUUCGCCACAUGUUUGUGUGUGCCAGCAGCACCACCCCACCGAAAGACCCCCACUCGACAACC ..((((.(((..(((((.(((((((.(((((((............)).))))).....(((((((....))))).)).))))))).)))))...(....).........))).)))) (-22.60 = -24.85 + 2.25)

| Location | 17,118,328 – 17,118,436 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.95 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17118328 108 - 22407834 GGUUGUCGAGUGGGGGUCUUUCGGUGGGGUGGUGCUGCAGGCGCACACAAACAUGUGGCGAAAACUAAUAGCAAAAAAGAGGAAGCAC---------CAGCACCCAGCAAAAACCGA ((((...(((.........)))(.((((((((((((.(...(((.((((....)))))))....((.............))).)))))---------)).).)))).)...)))).. ( -29.92) >DroSec_CAF1 82173 117 - 1 GGUUGUCGAGUGGGGGUCUUUCGGUGGGGUGGUGAUGCUGGCACACACAAACAUGUGGCGAAAACUAAUAGCAAAAAAGAGGAAGCACCAGUAGCAUCAGCACCCAGCAAAAACCGA ((((..((((.........))))((.(((((.((((((((((.((((......)))))).....................((.....))..)))))))).))))).))...)))).. ( -37.00) >DroSim_CAF1 98236 117 - 1 GGUUGUCGAGUGGGGGUCUUUCGGUGGGGUGGUGCUGCUGGCACACACAAACAUGUCGCGAAAACUAAUAACAAAAAAGAGGAAGCACUCGAAGCAGCAGCACCCAGCAAAAACCAA ((((..((((.........))))((.(((((.(((((((.(((((........))).))...................(((......)))..))))))).))))).))...)))).. ( -37.20) >DroEre_CAF1 84217 115 - 1 GCUUGUAGA--GUGGGGCUUUCGGUGGGGUGUUGCUGCUGGCACACACAAACAAGCGGCGAAAACUAAUAGCAAAAAAGAGGAAGCACCAGUAGCAUCAGCACCCAGCAAAAAGCGA .........--.....(((((..((.(((((((((((((((................((...........))........(....).)))))))))...)))))).))..))))).. ( -33.80) >consensus GGUUGUCGAGUGGGGGUCUUUCGGUGGGGUGGUGCUGCUGGCACACACAAACAUGUGGCGAAAACUAAUAGCAAAAAAGAGGAAGCACCAGUAGCAUCAGCACCCAGCAAAAACCGA ((((..((((.((...)).))))((.(((((.(((((((.((.(((........))))).....................(....)......))))))).))))).))...)))).. (-24.33 = -25.95 + 1.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:29 2006