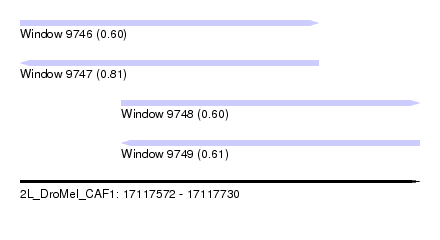
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,117,572 – 17,117,730 |
| Length | 158 |
| Max. P | 0.808955 |

| Location | 17,117,572 – 17,117,690 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17117572 118 + 22407834 CCUAAGCGCAUUAGCUUUAGUUUCUAGAAUUUUUUCUUAGCUUCCACUUGGCCGGCGAGGCCAAAGUUGGUUUUACCAAAGGAGCAAAGUGGUAAAAAAUGGAAAAAAAAAAUUGGCA .((((((......)).))))((((((...(((((.(((.(((((...((((((.....))))))..((((.....)))).))))).)))....))))).))))))............. ( -29.20) >DroSec_CAF1 81361 118 + 1 CCUAAGCGCAUUAGCUUUAGUUUCUAGCUUUUUUUCUUAGCUUCCACUUGGCCAGCUAGGCCAAAGUUGGUUUUACCAGAAGAGCAAAGUGGUAAAAAAAAUGAAAAAAAAAUUGGCA .((((((......)).))))......((((((((((.......((((((((((.....)))).))).)))((((((((...........)))))))).....))))))).....))). ( -27.00) >DroEre_CAF1 83444 115 + 1 ACUAAGCGCAUUAGCCUUAGUUUCUAGCAUUUUUUCGCAGCUUCCACUUGGCCAGCGAGGCCAAAGUUGGUCUAACCAGAAGAGCAAAGUGGUAAAAAGGGU---AAAAAAAUUGGCA ((((((.((....)))))))).....(((((((((..(..(((((((((((((.....))))...(((..(((....)))..))).))))))....))).).---.)))))))..)). ( -31.60) >DroYak_CAF1 83297 113 + 1 ACUAAGCGAAUUAGCUUCAGGUUCUAGCAUUUUUUUGAAGCUUCCACUUGGCCAGCGAGGCCAAAGUUGGUUUUACAAGAAGAGCAAAGUGGUAAAAAAG-U----ACAAAAUUGCAA .....((((...(((((((((............))))))))).((((((((((.....))))))..(((.((((.....)))).))).))))........-.----......)))).. ( -29.00) >consensus ACUAAGCGCAUUAGCUUUAGUUUCUAGCAUUUUUUCGUAGCUUCCACUUGGCCAGCGAGGCCAAAGUUGGUUUUACCAGAAGAGCAAAGUGGUAAAAAAGGU___AAAAAAAUUGGCA .((((((......)).)))).......................((((((((((.....))))))..(((.((((.....)))).))).)))).......................... (-20.33 = -20.45 + 0.12)

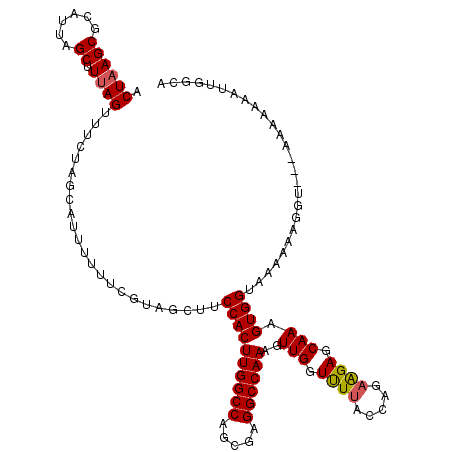

| Location | 17,117,572 – 17,117,690 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17117572 118 - 22407834 UGCCAAUUUUUUUUUUCCAUUUUUUACCACUUUGCUCCUUUGGUAAAACCAACUUUGGCCUCGCCGGCCAAGUGGAAGCUAAGAAAAAAUUCUAGAAACUAAAGCUAAUGCGCUUAGG ....((((((((((((((((.((((((((...........))))))))......((((((.....)))))))))))....))))))))))).......((((.((....))..)))). ( -28.30) >DroSec_CAF1 81361 118 - 1 UGCCAAUUUUUUUUUCAUUUUUUUUACCACUUUGCUCUUCUGGUAAAACCAACUUUGGCCUAGCUGGCCAAGUGGAAGCUAAGAAAAAAAGCUAGAAACUAAAGCUAAUGCGCUUAGG .......(((((((((.....((((((((...........))))))))(((...((((((.....)))))).))).......))))))))).......((((.((....))..)))). ( -29.20) >DroEre_CAF1 83444 115 - 1 UGCCAAUUUUUUU---ACCCUUUUUACCACUUUGCUCUUCUGGUUAGACCAACUUUGGCCUCGCUGGCCAAGUGGAAGCUGCGAAAAAAUGCUAGAAACUAAGGCUAAUGCGCUUAGU .............---.....(((((.((.(((((.(((((((.....)).((((.((((.....)))))))))))))..)))))....)).)))))((((((((....)).)))))) ( -29.10) >DroYak_CAF1 83297 113 - 1 UUGCAAUUUUGU----A-CUUUUUUACCACUUUGCUCUUCUUGUAAAACCAACUUUGGCCUCGCUGGCCAAGUGGAAGCUUCAAAAAAAUGCUAGAACCUGAAGCUAAUUCGCUUAGU ..((........----.-........(((((((((.......))))).......((((((.....)))))))))).(((((((................))))))).....))..... ( -26.69) >consensus UGCCAAUUUUUUU___ACCUUUUUUACCACUUUGCUCUUCUGGUAAAACCAACUUUGGCCUCGCUGGCCAAGUGGAAGCUAAGAAAAAAUGCUAGAAACUAAAGCUAAUGCGCUUAGG .....((((((((........((((((((...........))))))))(((...((((((.....)))))).))).......))))))))........(((.(((......)))))). (-20.56 = -21.25 + 0.69)

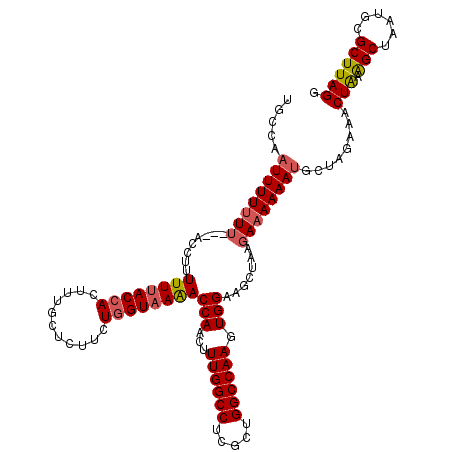

| Location | 17,117,612 – 17,117,730 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -31.73 |
| Energy contribution | -31.47 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17117612 118 + 22407834 CUUCCACUUGGCCGGCGAGGCCAAAGUUGGUUUUACCAAAGGAGCAAAGUGGUAAAAAAUGGAAAAAAAAAAUUGGCAGAAGGUCGCUUCUCUGCAUUAGCUGGUCUGCAAGCGGCUC .(((((.((((((.....))))))......((((((((...........))))))))..)))))...........(((((.((.....)))))))...(((((.(.....).))))). ( -32.00) >DroSec_CAF1 81401 118 + 1 CUUCCACUUGGCCAGCUAGGCCAAAGUUGGUUUUACCAGAAGAGCAAAGUGGUAAAAAAAAUGAAAAAAAAAUUGGCAGAAGGUCGCUUCUCUGCAUUAGCUGGCCUGCAAGCGGCUC ...((.(((((((((((((((((...(((.((((.....)))).)))..))))......................(((((.((.....))))))).))))))))))...))).))... ( -36.90) >DroEre_CAF1 83484 115 + 1 CUUCCACUUGGCCAGCGAGGCCAAAGUUGGUCUAACCAGAAGAGCAAAGUGGUAAAAAGGGU---AAAAAAAUUGGCAGAAGGUCGCUCCUCUGCAUUAGCUGGUCUGCAAGCGGCUC ...((.((((((((((...((((...(((.(((.......))).)))..)))).........---..........(((((.((.....)))))))....))))))...)))).))... ( -35.60) >DroYak_CAF1 83337 113 + 1 CUUCCACUUGGCCAGCGAGGCCAAAGUUGGUUUUACAAGAAGAGCAAAGUGGUAAAAAAG-U----ACAAAAUUGCAAGAAGGUCGCUCCUCUGCAUUAGCUGGUCCGCAAGCGGCUC ...((.((((((((((...((((...(((.((((.....)))).)))..)))).......-.----.......((((.((.((.....))))))))...))))))...)))).))... ( -31.20) >consensus CUUCCACUUGGCCAGCGAGGCCAAAGUUGGUUUUACCAGAAGAGCAAAGUGGUAAAAAAGGU___AAAAAAAUUGGCAGAAGGUCGCUCCUCUGCAUUAGCUGGUCUGCAAGCGGCUC ...((.((((((((((...((((...(((.((((.....)))).)))..))))......................(((((.((.....)))))))....))))))...)))).))... (-31.73 = -31.47 + -0.25)

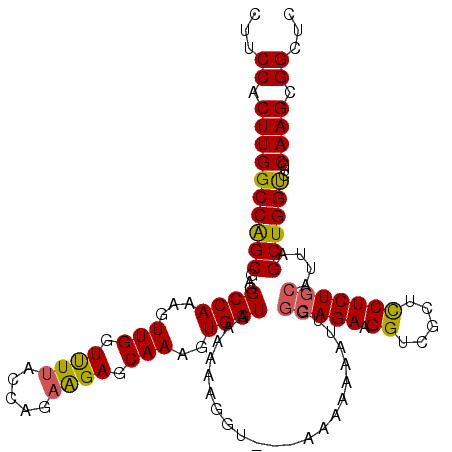
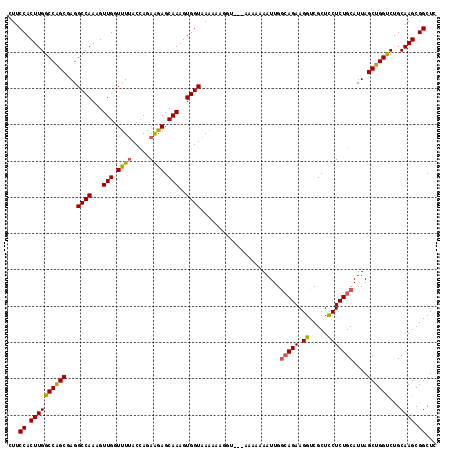
| Location | 17,117,612 – 17,117,730 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.79 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.29 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17117612 118 - 22407834 GAGCCGCUUGCAGACCAGCUAAUGCAGAGAAGCGACCUUCUGCCAAUUUUUUUUUUCCAUUUUUUACCACUUUGCUCCUUUGGUAAAACCAACUUUGGCCUCGCCGGCCAAGUGGAAG .....(((........)))....((((((........))))))...........((((((.((((((((...........))))))))......((((((.....)))))))))))). ( -28.40) >DroSec_CAF1 81401 118 - 1 GAGCCGCUUGCAGGCCAGCUAAUGCAGAGAAGCGACCUUCUGCCAAUUUUUUUUUCAUUUUUUUUACCACUUUGCUCUUCUGGUAAAACCAACUUUGGCCUAGCUGGCCAAGUGGAAG ...((((((...(((((((((..((((((........))))))..................((((((((...........))))))))(((....)))..)))))))))))))))... ( -36.90) >DroEre_CAF1 83484 115 - 1 GAGCCGCUUGCAGACCAGCUAAUGCAGAGGAGCGACCUUCUGCCAAUUUUUUU---ACCCUUUUUACCACUUUGCUCUUCUGGUUAGACCAACUUUGGCCUCGCUGGCCAAGUGGAAG ...(((((((..(.(((((......((((((((((..................---...............))))))))))(((((((.....)))))))..)))))))))))))... ( -33.91) >DroYak_CAF1 83337 113 - 1 GAGCCGCUUGCGGACCAGCUAAUGCAGAGGAGCGACCUUCUUGCAAUUUUGU----A-CUUUUUUACCACUUUGCUCUUCUUGUAAAACCAACUUUGGCCUCGCUGGCCAAGUGGAAG ...((((((..((.(((((...((((((((((((.....).((((....)))----)-...............))))))).))))...(((....)))....)))))))))))))... ( -33.00) >consensus GAGCCGCUUGCAGACCAGCUAAUGCAGAGAAGCGACCUUCUGCCAAUUUUUUU___ACCUUUUUUACCACUUUGCUCUUCUGGUAAAACCAACUUUGGCCUCGCUGGCCAAGUGGAAG ...(((((((..(.(((((.....(((((((((((....................................)))))))))))......(((....)))....)))))))))))))... (-27.67 = -28.29 + 0.62)

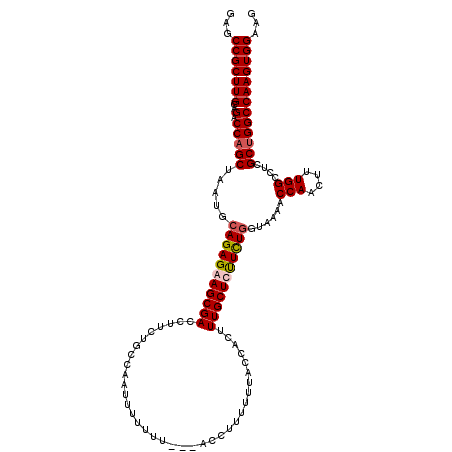

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:24 2006