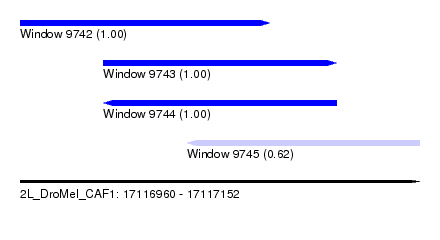
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,116,960 – 17,117,152 |
| Length | 192 |
| Max. P | 0.999958 |

| Location | 17,116,960 – 17,117,080 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.92 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -36.63 |
| Energy contribution | -36.67 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17116960 120 + 22407834 GCGCAAGGGAAAUGCUCCAAAAAUACCUAAAUGCGUUAAUUGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGAAACGCCAAUGGCGGAUGCGUUGGCAGCGGAUG ((((..((..(((((.................)))))..))..))))..(((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))) ( -37.83) >DroSec_CAF1 80763 120 + 1 GUGCAAGGGAAAUGCUCCAAAAAUACCUAACUGCGUUAAUUGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGUGGCGGAUGCGUUGGCAGCGGAUG ((((..((..(((((.................)))))..))..))))..(((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))) ( -35.93) >DroSim_CAF1 96433 120 + 1 GCGCAAGGGAAAUGCUCCAAAAAUACCUAACUGCGUUAAUUGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGUGGCGGAUGCGUUGGCAGCGGAUG ((((..((..(((((.................)))))..))..))))..(((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))) ( -37.83) >DroEre_CAF1 82859 120 + 1 GCGCAGGGGAAAUGCUCCAAAAAUACCUAACUGCGUUAAUUGAGCGCGACAUUUGCUGAAACUAGAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGGGGCGGAUGCGUUGGCAGCGGAUG ((((.((((.....)))).........((((...)))).....))))..(((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))) ( -38.70) >DroYak_CAF1 82710 120 + 1 GCGCAGGGGAAAUGCUCCAAAAAUACCUAACUGCGUUAAUUGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGAGGCGGAUGCGUUGGCAGCGGAUA ((((.((((.....)))).........((((...)))).....))))...((((((((.(((....(((((....)))))...(((....((((...))))..))))))..)))))))). ( -36.60) >consensus GCGCAAGGGAAAUGCUCCAAAAAUACCUAACUGCGUUAAUUGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGUGGCGGAUGCGUUGGCAGCGGAUG ((((..((..(((((.................)))))..))..))))..(((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))) (-36.63 = -36.67 + 0.04)

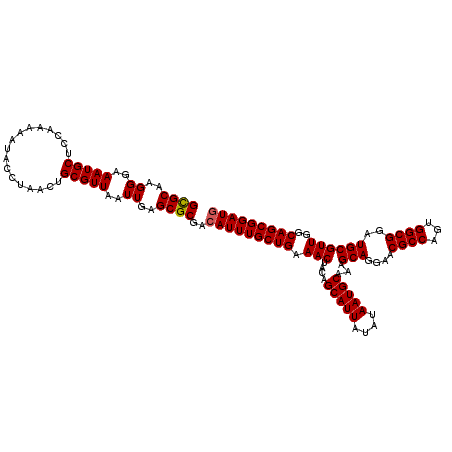
| Location | 17,117,000 – 17,117,112 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -39.76 |
| Consensus MFE | -36.90 |
| Energy contribution | -37.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.87 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
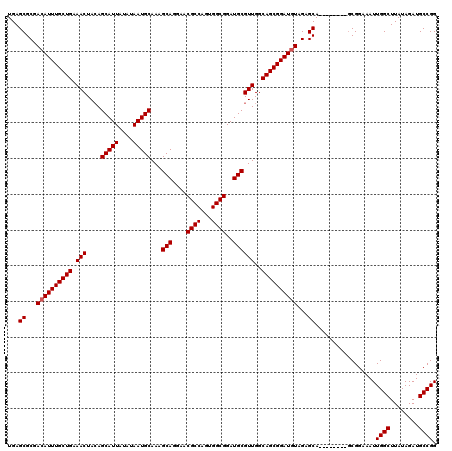
>2L_DroMel_CAF1 17117000 112 + 22407834 UGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGAAACGCCAAUGGCGGAUGCGUUGGCAGCGGAUGUAGAGCA--------GCGGAAAUUGGCUUAUAGAUGCCGG ...((((.((((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))))...)).--------))......((((........)))). ( -39.70) >DroSec_CAF1 80803 112 + 1 UGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGUGGCGGAUGCGUUGGCAGCGGAUGUAGAGCA--------GCGGAAAUUGGCUUAUAGAUGCCGG ...((((.((((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))))...)).--------))......((((........)))). ( -39.70) >DroSim_CAF1 96473 112 + 1 UGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGUGGCGGAUGCGUUGGCAGCGGAUGUAGAGCA--------UCGGAAAUUGGCUUAUAGAUGCCGG .((((...((((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))))...)).--------))......((((........)))). ( -38.90) >DroEre_CAF1 82899 120 + 1 UGAGCGCGACAUUUGCUGAAACUAGAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGGGGCGGAUGCGUUGGCAGCGGAUGUAGCGCAGAAAUAGCGCGGAAAUUGGCUUAUAGAUGCCGG ...((((.((((((((((.(((....(((((....)))))...(((....((((...))))..))))))..)))))))))).)))).................((((........)))). ( -43.60) >DroYak_CAF1 82750 112 + 1 UGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGAGGCGGAUGCGUUGGCAGCGGAUAUAGUGCA--------GCGGAAAUUGGCUUAUAGAUGCCGG ...((((...((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))...)))).--------........((((........)))). ( -36.90) >consensus UGAGCGCGACAUUUGCUGAAACUACAGCAUUAUAUAAUGCAAAGCAGGAACGCCAGUGGCGGAUGCGUUGGCAGCGGAUGUAGAGCA________GCGGAAAUUGGCUUAUAGAUGCCGG ...((...((((((((((.(((....(((((....)))))...(((....((((...))))..))))))..))))))))))...)).................((((........)))). (-36.90 = -37.10 + 0.20)

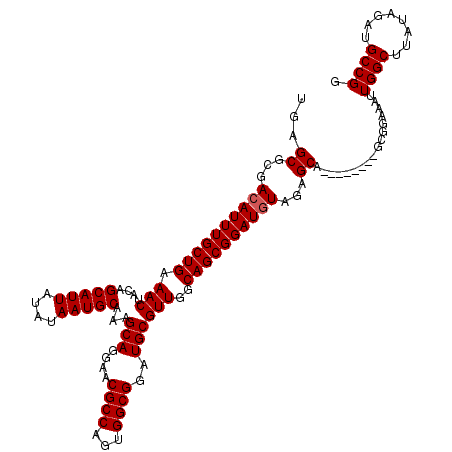
| Location | 17,117,000 – 17,117,112 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
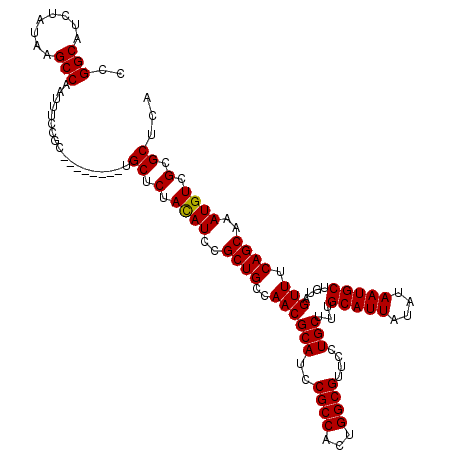
>2L_DroMel_CAF1 17117000 112 - 22407834 CCGGCAUCUAUAAGCCAAUUUCCGC--------UGCUCUACAUCCGCUGCCAACGCAUCCGCCAUUGGCGUUUCUGCUUUGCAUUAUAUAAUGCUGUAGUUUCAGCAAAUGUCGCGCUCA ..(((........))).........--------.((.(.((((..((((((((.((....))..)))))(...((((...(((((....))))).))))...))))..)))).).))... ( -28.80) >DroSec_CAF1 80803 112 - 1 CCGGCAUCUAUAAGCCAAUUUCCGC--------UGCUCUACAUCCGCUGCCAACGCAUCCGCCACUGGCGUUCCUGCUUUGCAUUAUAUAAUGCUGUAGUUUCAGCAAAUGUCGCGCUCA ..(((........))).........--------.((.(.((((..(((((((..((....))...))))....((((...(((((....))))).))))....)))..)))).).))... ( -27.40) >DroSim_CAF1 96473 112 - 1 CCGGCAUCUAUAAGCCAAUUUCCGA--------UGCUCUACAUCCGCUGCCAACGCAUCCGCCACUGGCGUUCCUGCUUUGCAUUAUAUAAUGCUGUAGUUUCAGCAAAUGUCGCGCUCA ..(((........))).......((--------.((.(.((((..(((((((..((....))...))))....((((...(((((....))))).))))....)))..)))).).)))). ( -28.90) >DroEre_CAF1 82899 120 - 1 CCGGCAUCUAUAAGCCAAUUUCCGCGCUAUUUCUGCGCUACAUCCGCUGCCAACGCAUCCGCCCCUGGCGUUCCUGCUUUGCAUUAUAUAAUGCUCUAGUUUCAGCAAAUGUCGCGCUCA ..(((........)))..................((((.((((..((((..((((((..((((...))))....)))...(((((....)))))....))).))))..)))).))))... ( -32.70) >DroYak_CAF1 82750 112 - 1 CCGGCAUCUAUAAGCCAAUUUCCGC--------UGCACUAUAUCCGCUGCCAACGCAUCCGCCUCUGGCGUUCCUGCUUUGCAUUAUAUAAUGCUGUAGUUUCAGCAAAUGUCGCGCUCA ..(((........)))......(((--------.(((.(((((....(((.((.(((..((((...))))....))).)))))..))))).(((((......)))))..))).))).... ( -26.30) >consensus CCGGCAUCUAUAAGCCAAUUUCCGC________UGCUCUACAUCCGCUGCCAACGCAUCCGCCACUGGCGUUCCUGCUUUGCAUUAUAUAAUGCUGUAGUUUCAGCAAAUGUCGCGCUCA ..(((........)))..................((.(.((((..((((..((((((..((((...))))....)))...(((((....)))))....))).))))..)))).).))... (-26.68 = -26.52 + -0.16)

| Location | 17,117,040 – 17,117,152 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
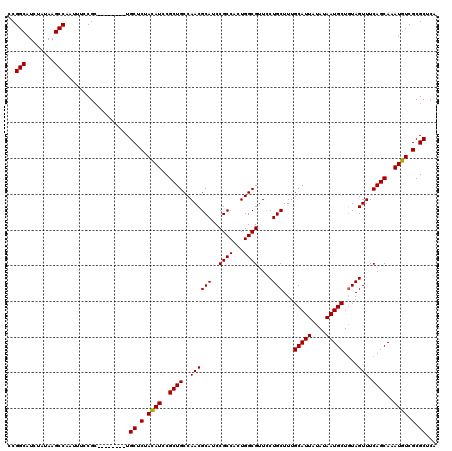
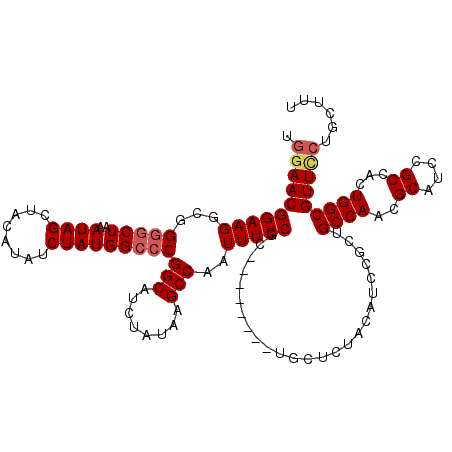
>2L_DroMel_CAF1 17117040 112 - 22407834 UGGAACGGAAGGCGGGGCUAAAUAGCUACAUAUCUAUGGCCCGGCAUCUAUAAGCCAAUUUCCGC--------UGCUCUACAUCCGCUGCCAACGCAUCCGCCAUUGGCGUUUCUGCUUU .((((((((.(((((((((..((((........)))))))))(((........)))......)))--------)..))).........(((((.((....))..))))))))))...... ( -34.00) >DroSec_CAF1 80843 112 - 1 UGGAACGGAAGGCGGGGCUAAAUAGCUACAUAUCUAUGGCCCGGCAUCUAUAAGCCAAUUUCCGC--------UGCUCUACAUCCGCUGCCAACGCAUCCGCCACUGGCGUUCCUGCUUU .((((((((.(((((((((..((((........)))))))))(((........)))......)))--------)..))).........((((..((....))...)))))))))...... ( -36.00) >DroSim_CAF1 96513 112 - 1 UGGAACGGAAGGCGGGGCUAAAUAGCUACAUAUCUAUGGCCCGGCAUCUAUAAGCCAAUUUCCGA--------UGCUCUACAUCCGCUGCCAACGCAUCCGCCACUGGCGUUCCUGCUUU .((((((((.(((((((((..((((........)))))))))(((........))).......((--------((.....))))))))((....)).)))(((...))))))))...... ( -35.70) >DroEre_CAF1 82939 117 - 1 UCGAACGGAAGACGG--CU-AAUAGCUGCAUAUCUAUGGCCCGGCAUCUAUAAGCCAAUUUCCGCGCUAUUUCUGCGCUACAUCCGCUGCCAACGCAUCCGCCCCUGGCGUUCCUGCUUU ..(((((((((..((--((-.((((.(((....(....)....))).)))).))))..)))))((((.......))))..........((((..((....))...))))))))....... ( -35.00) >DroYak_CAF1 82790 112 - 1 UGAAACGGAAGGCGGGGCUAAAUAGCUACAUAUCUAUGGCCCGGCAUCUAUAAGCCAAUUUCCGC--------UGCACUAUAUCCGCUGCCAACGCAUCCGCCUCUGGCGUUCCUGCUUU ......(((((((((((((..((((........)))))))))(((........)))......)))--------)..............((((..((....))...)))).))))...... ( -30.20) >consensus UGGAACGGAAGGCGGGGCUAAAUAGCUACAUAUCUAUGGCCCGGCAUCUAUAAGCCAAUUUCCGC________UGCUCUACAUCCGCUGCCAACGCAUCCGCCACUGGCGUUCCUGCUUU .((((((((((...(((((..((((........)))))))))(((........)))..))))).........................((((..((....))...)))))))))...... (-28.00 = -28.68 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:20 2006