| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,116,287 – 17,116,400 |
| Length | 113 |
| Max. P | 0.985229 |

| Location | 17,116,287 – 17,116,400 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17116287 113 + 22407834 AACAACAGGAAAACCAAAAGAGUGAAAGAGCGAA-AAAAAGUUUUGAAAUGAAAAAGCAGUCAGCAUAUGAAAUGAGUUUAUUACUCAAAAUGCUCCGCUCCGUGUUUAU------AUGU ...((((((....))....(((((...(((((..-.....(((((........)))))..(((.....)))..(((((.....)))))...))))))))))..))))...------.... ( -20.50) >DroSec_CAF1 80090 114 + 1 AACAACAGGAAAACCAAAAGAGUGAAAGAGCGAGAAAAAAGUUUUGAAAUGAAAAAGCAGUCAGCAUAUGAAAUGAGUUUAUUACUCAAAAUGCUCCGCUCCGUGUUUAU------AUGU ...((((((....))....(((((...(((((........(((((........)))))..(((.....)))..(((((.....)))))...))))))))))..))))...------.... ( -20.50) >DroSim_CAF1 95752 114 + 1 AACAACAGGAAAACCAAAAGAGUGAAAGAGCGAAAAAAAAGUUUUGAAAUGAAAAAGCAGUCAGCAUAUGAAAUGAGUUUAUUACUCAAAAUGCUCCGCUCCGUGUUUAU------AUGU ...((((((....))....(((((...(((((........(((((........)))))..(((.....)))..(((((.....)))))...))))))))))..))))...------.... ( -20.50) >DroEre_CAF1 82187 120 + 1 AACAACAGGAAAACCAAAACAGUGAAAGAGCGAAAAAAAAGUUUUGAAAUGAAAAAGCAGUCAGCAUAUGAAAUGAGUUUAUUACUCAAAAUGCUCCACUCCGUGUUUAUACGAGCAUGU .......((....)).....((((...(((((........(((((........)))))..(((.....)))..(((((.....)))))...))))))))).(((((((....))))))). ( -21.30) >DroYak_CAF1 82138 116 + 1 AACAACAGGAAAACCAAUAGAGUGAAAGAGCGAAA--AAAGUUUUGAAAUGAAAAAGCAGUCAGCAUAUGAAAUGAGUUUAUUACUCAAAAUGCUCCACUCGCUGUUUAUACA--UAUGU ((((.(.((....))....(((((...(((((...--...(((((........)))))..(((.....)))..(((((.....)))))...))))))))))).))))......--..... ( -21.90) >consensus AACAACAGGAAAACCAAAAGAGUGAAAGAGCGAAAAAAAAGUUUUGAAAUGAAAAAGCAGUCAGCAUAUGAAAUGAGUUUAUUACUCAAAAUGCUCCGCUCCGUGUUUAU______AUGU ...((((((....))....(((((...(((((........(((((........)))))..(((.....)))..(((((.....)))))...))))))))))..))))............. (-20.08 = -20.04 + -0.04)


| Location | 17,116,287 – 17,116,400 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -19.99 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17116287 113 - 22407834 ACAU------AUAAACACGGAGCGGAGCAUUUUGAGUAAUAAACUCAUUUCAUAUGCUGACUGCUUUUUCAUUUCAAAACUUUUU-UUCGCUCUUUCACUCUUUUGGUUUUCCUGUUGUU ....------...((((.((((((((((((..(((((.....)))))......))))(((........)))..............-))))))))...........((....))))))... ( -19.30) >DroSec_CAF1 80090 114 - 1 ACAU------AUAAACACGGAGCGGAGCAUUUUGAGUAAUAAACUCAUUUCAUAUGCUGACUGCUUUUUCAUUUCAAAACUUUUUUCUCGCUCUUUCACUCUUUUGGUUUUCCUGUUGUU ....------...((((.((((((((((((..(((((.....)))))......)))))((........)).................)))))))...........((....))))))... ( -18.60) >DroSim_CAF1 95752 114 - 1 ACAU------AUAAACACGGAGCGGAGCAUUUUGAGUAAUAAACUCAUUUCAUAUGCUGACUGCUUUUUCAUUUCAAAACUUUUUUUUCGCUCUUUCACUCUUUUGGUUUUCCUGUUGUU ....------...((((.((((((((((((..(((((.....)))))......))))(((........)))...............))))))))...........((....))))))... ( -19.30) >DroEre_CAF1 82187 120 - 1 ACAUGCUCGUAUAAACACGGAGUGGAGCAUUUUGAGUAAUAAACUCAUUUCAUAUGCUGACUGCUUUUUCAUUUCAAAACUUUUUUUUCGCUCUUUCACUGUUUUGGUUUUCCUGUUGUU .............((((.((((..((((((..(((((.....)))))......)))))..)..).........(((((((....................)))))))...)))))))... ( -19.75) >DroYak_CAF1 82138 116 - 1 ACAUA--UGUAUAAACAGCGAGUGGAGCAUUUUGAGUAAUAAACUCAUUUCAUAUGCUGACUGCUUUUUCAUUUCAAAACUUU--UUUCGCUCUUUCACUCUAUUGGUUUUCCUGUUGUU .....--......(((((((((((((((....(((((.....)))))........((.....))...................--....))))...)))))....((....)).)))))) ( -23.00) >consensus ACAU______AUAAACACGGAGCGGAGCAUUUUGAGUAAUAAACUCAUUUCAUAUGCUGACUGCUUUUUCAUUUCAAAACUUUUUUUUCGCUCUUUCACUCUUUUGGUUUUCCUGUUGUU .............((((.((((((((((((..(((((.....)))))......)))))((........)).................)))))))...........((....))))))... (-17.52 = -17.48 + -0.04)

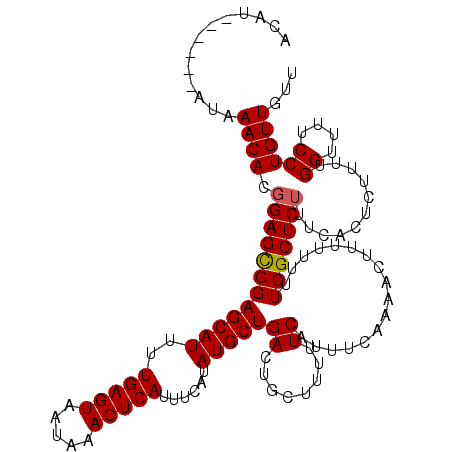
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:17 2006