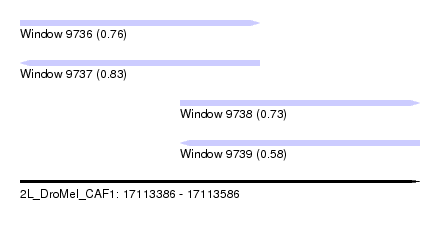
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,113,386 – 17,113,586 |
| Length | 200 |
| Max. P | 0.831875 |

| Location | 17,113,386 – 17,113,506 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17113386 120 + 22407834 AGAAAGCAGCGCAAACUGACAAAGUUUCUUGUGCUUUUCGCAACUUUCUUCUGCUCAUCCCGCCCACUUUUCCGCCUCCCUUCCCUUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCU .((((..((((((((((.....)))...)))))))))))(((.........)))..........................................((((((((.....)).)))))).. ( -19.20) >DroSec_CAF1 77393 120 + 1 AGAAAGCAGCGCAAACUGACAAAGUUUCUUGUGCUUUUCGCAACUUUCUUCUGCUCAUCCCGCCCACUUUUCCGCCGCCCUUCCCUUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCU .(((.((((((((((((.....)))...)))))))....(((.........)))......................))..))).............((((((((.....)).)))))).. ( -19.40) >DroSim_CAF1 91022 120 + 1 AGAAAGCAGCGCAAACUGACAAAGUUUCUUGUGCUUUUCGCAACUUUCUUCUGCUCAUCCCGCCCACUUUUCCGCCGCCCAUCCCGUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCU .((((((((((((((((.....)))...)))))))....(((.........)))...............................)))))).....((((((((.....)).)))))).. ( -22.50) >DroEre_CAF1 78782 120 + 1 AGAAAGCAGCGCAAACUGACAAAGUUUCUUGUGCUUUUCGCAACUUUCCCCUGCUCAUCCCGCCCACUUUUCCGCCUCCCUCCUCUUUUUCCUUUUGCGAACGCCUUUUGCUGUUCGCCU .((((..((((((((((.....)))...)))))))))))(((.........)))..........................................((((((((.....)).)))))).. ( -21.00) >DroYak_CAF1 79249 120 + 1 AGAAAGCCUUGCAAACUGACAAAGUUUCUUUUGCUUUUCGCAACUUUCUCCUGCUCAUCCCACCUUCUUUUCCACUUCCCUUCUCACUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCU ((((((..((((.((..(.(((((....))))))..)).))))))))))...............................................((((((((.....)).)))))).. ( -16.30) >consensus AGAAAGCAGCGCAAACUGACAAAGUUUCUUGUGCUUUUCGCAACUUUCUUCUGCUCAUCCCGCCCACUUUUCCGCCUCCCUUCCCUUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCU .(((((((....(((((.....)))))....))))))).(((.........)))..........................................((((((((.....)).)))))).. (-17.40 = -17.44 + 0.04)

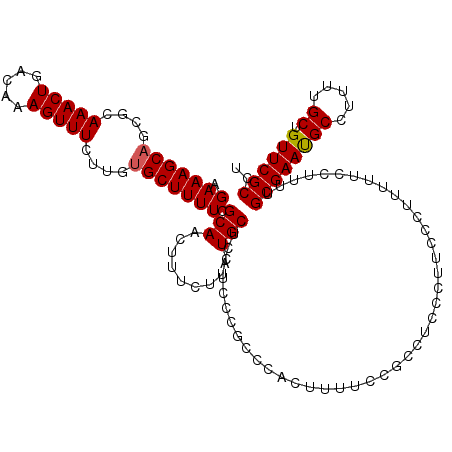
| Location | 17,113,386 – 17,113,506 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17113386 120 - 22407834 AGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAAAGGGAAGGGAGGCGGAAAAGUGGGCGGGAUGAGCAGAAGAAAGUUGCGAAAAGCACAAGAAACUUUGUCAGUUUGCGCUGCUUUCU ..(((((..((.....)).)))))................(((((((((....(..(((..((..((...........(((.....)))........))..)).)))..).))))))))) ( -28.31) >DroSec_CAF1 77393 120 - 1 AGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAAAGGGAAGGGCGGCGGAAAAGUGGGCGGGAUGAGCAGAAGAAAGUUGCGAAAAGCACAAGAAACUUUGUCAGUUUGCGCUGCUUUCU ..(((((..((.....)).)))))............((((((.(((((.(((.....((.......)).((..((((((((.....)))......))))).))..))).))))))))))) ( -28.90) >DroSim_CAF1 91022 120 - 1 AGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAACGGGAUGGGCGGCGGAAAAGUGGGCGGGAUGAGCAGAAGAAAGUUGCGAAAAGCACAAGAAACUUUGUCAGUUUGCGCUGCUUUCU ..(((((..((.....)).)))))............(((.((((((((.(((.....((.......)).((..((((((((.....)))......))))).))..))).))))))))))) ( -28.50) >DroEre_CAF1 78782 120 - 1 AGGCGAACAGCAAAAGGCGUUCGCAAAAGGAAAAAGAGGAGGGAGGCGGAAAAGUGGGCGGGAUGAGCAGGGGAAAGUUGCGAAAAGCACAAGAAACUUUGUCAGUUUGCGCUGCUUUCU ..((((((.((.....))))))))................(((((((((....(..(((....(((.(((((......(((.....))).......)))))))))))..).))))))))) ( -34.22) >DroYak_CAF1 79249 120 - 1 AGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAGUGAGAAGGGAAGUGGAAAAGAAGGUGGGAUGAGCAGGAGAAAGUUGCGAAAAGCAAAAGAAACUUUGUCAGUUUGCAAGGCUUUCU ..(((((..((.....)).)))))....(((((((.....(.(((.((..((((...((.......)).........((((.....))))......))))..)).))).)...))))))) ( -22.90) >consensus AGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAAAGGGAAGGGAGGCGGAAAAGUGGGCGGGAUGAGCAGAAGAAAGUUGCGAAAAGCACAAGAAACUUUGUCAGUUUGCGCUGCUUUCU ..(((((..((.....)).)))))................(((((((((....(..(((..((..(..((........(((.....))).......)))..)).)))..).))))))))) (-23.38 = -23.70 + 0.32)

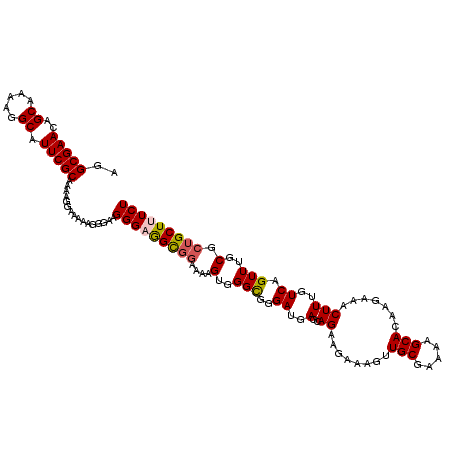
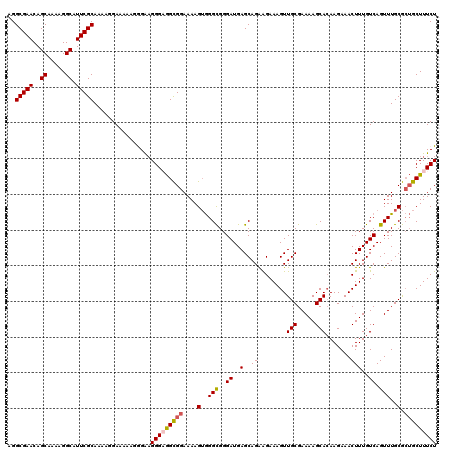
| Location | 17,113,466 – 17,113,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17113466 120 + 22407834 UUCCCUUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCUUGGUUUUCCUAUUAUUUGCGCAUUUUAUUUCCAUUGAACAUUUCAUUGACACUUUGACAGCAGCAUUGACAUUUGCGUGA ................(((((((.(...((((((((((..((((......))))...)))...........((.(((.....))).))..........)))))))..).))))))).... ( -21.80) >DroSec_CAF1 77473 120 + 1 UUCCCUUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCUUGGUUUUCCUAUUAUUUGCGCAUUUUAUUUCCAUUGAACAUUUCAUUGACACUUUGACAGCAGCAUUGACAUUUGCGUGA ................(((((((.(...((((((((((..((((......))))...)))...........((.(((.....))).))..........)))))))..).))))))).... ( -21.80) >DroSim_CAF1 91102 120 + 1 AUCCCGUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCUUGGUUUUCCUAUUAUUUGCGCAUUUUAUUUCCAUUGAACAUUUCAUUGACACUUUGACAGCAGCAUUGACAUUUGCGUGA ................(((((((.(...((((((((((..((((......))))...)))...........((.(((.....))).))..........)))))))..).))))))).... ( -21.80) >DroEre_CAF1 78862 120 + 1 UCCUCUUUUUCCUUUUGCGAACGCCUUUUGCUGUUCGCCUUGCUUCUCCUAUUAUUUGCGCAUUUUAUUUCCAUUGAACAUUUCAUUGACACUUUGACAGCAGCAUUGACAUUUGCGUGA ................((((((((.....)).)))))).................(..((((.........((.(((.....))).))...(..((.(....)))..).....))))..) ( -17.10) >DroYak_CAF1 79329 120 + 1 UUCUCACUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCUUGGUCCUUCUAUUAUUUGCGCAUUUUAUUUCCAUUGAACAUUUCAUUGACACUUUGACAGCAGCAUUGACAUUUGCGUGA ................(((((((.(...((((((((((..((((......))))...)))...........((.(((.....))).))..........)))))))..).))))))).... ( -21.00) >consensus UUCCCUUUUUCCUUUUGCGAAUGCCUUUUGCUGUUCGCCUUGGUUUUCCUAUUAUUUGCGCAUUUUAUUUCCAUUGAACAUUUCAUUGACACUUUGACAGCAGCAUUGACAUUUGCGUGA ................(((((((.(...((((((((((..((((......))))...)))...........((.(((.....))).))..........)))))))..).))))))).... (-20.22 = -20.62 + 0.40)

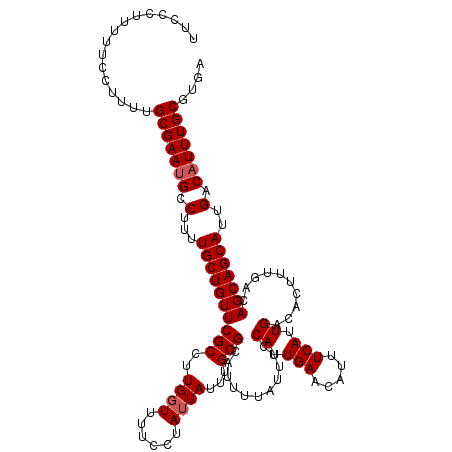
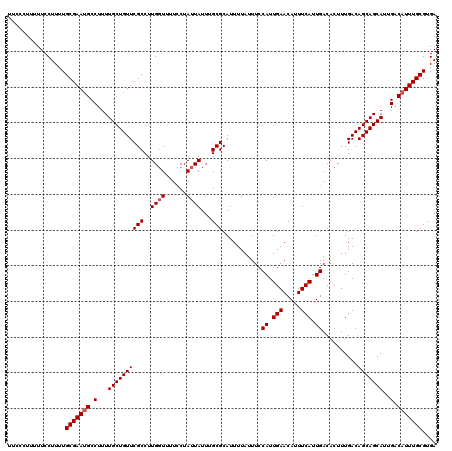
| Location | 17,113,466 – 17,113,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -20.13 |
| Energy contribution | -19.97 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17113466 120 - 22407834 UCACGCAAAUGUCAAUGCUGCUGUCAAAGUGUCAAUGAAAUGUUCAAUGGAAAUAAAAUGCGCAAAUAAUAGGAAAACCAAGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAAAGGGAA ....((.((((((..(((((.((((...((((..........(((....))).......))))........((....))..))))..)))))...)))))).))................ ( -21.93) >DroSec_CAF1 77473 120 - 1 UCACGCAAAUGUCAAUGCUGCUGUCAAAGUGUCAAUGAAAUGUUCAAUGGAAAUAAAAUGCGCAAAUAAUAGGAAAACCAAGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAAAGGGAA ....((.((((((..(((((.((((...((((..........(((....))).......))))........((....))..))))..)))))...)))))).))................ ( -21.93) >DroSim_CAF1 91102 120 - 1 UCACGCAAAUGUCAAUGCUGCUGUCAAAGUGUCAAUGAAAUGUUCAAUGGAAAUAAAAUGCGCAAAUAAUAGGAAAACCAAGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAACGGGAU ....((.((((((..(((((.((((...((((..........(((....))).......))))........((....))..))))..)))))...)))))).))................ ( -21.93) >DroEre_CAF1 78862 120 - 1 UCACGCAAAUGUCAAUGCUGCUGUCAAAGUGUCAAUGAAAUGUUCAAUGGAAAUAAAAUGCGCAAAUAAUAGGAGAAGCAAGGCGAACAGCAAAAGGCGUUCGCAAAAGGAAAAAGAGGA ....((.((((((..(((((.((((......(((.(((.....))).)))........(((................))).))))..)))))...)))))).))................ ( -21.19) >DroYak_CAF1 79329 120 - 1 UCACGCAAAUGUCAAUGCUGCUGUCAAAGUGUCAAUGAAAUGUUCAAUGGAAAUAAAAUGCGCAAAUAAUAGAAGGACCAAGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAGUGAGAA ((((.(...(((.(((((((((((.......(((.(((.....))).)))..........(((...................))).)))))....)))))).)))....)...))))... ( -20.61) >consensus UCACGCAAAUGUCAAUGCUGCUGUCAAAGUGUCAAUGAAAUGUUCAAUGGAAAUAAAAUGCGCAAAUAAUAGGAAAACCAAGGCGAACAGCAAAAGGCAUUCGCAAAAGGAAAAAGGGAA ....((.((((((..(((((.((((...((((..........(((....))).......))))........(......)..))))..)))))...)))))).))................ (-20.13 = -19.97 + -0.16)

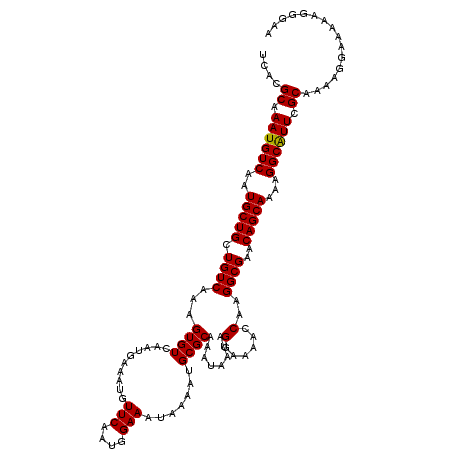
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:15 2006