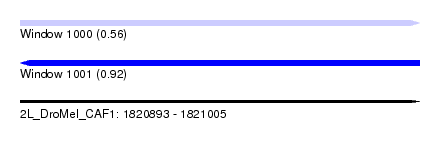
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,820,893 – 1,821,005 |
| Length | 112 |
| Max. P | 0.922666 |

| Location | 1,820,893 – 1,821,005 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
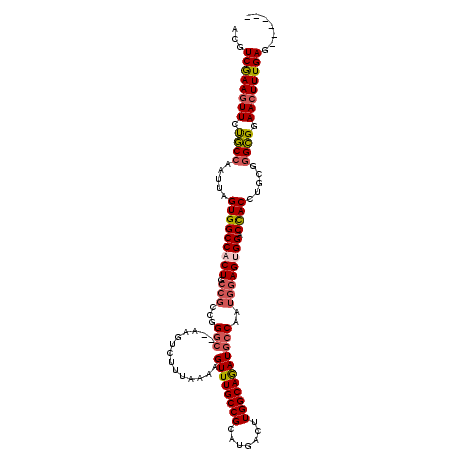
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -30.23 |
| Energy contribution | -31.37 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1820893 112 + 22407834 ------CUCAAAGUUCCGCCCGCAGGUGGUCCACUCCAUUGGCAUCUGCCAAGUCAUGCGGCAAACUUUUAAAGACUU--GCCCGGCGGCAGUGGCCACUAAUUGGCAGAACUUCGACGU ------.((.((((((.(((....(((((.(((((((.(((((....)))))(((..(.(((((............))--))))))))).))))))))))....))).)))))).))... ( -44.10) >DroSec_CAF1 40907 112 + 1 ------CUCAAAGUUCCGCCCGCCGGUGGUCCACUCCAUUGGCAUCUGCCAAGUCAUGCGGCAAACUUUUAAAGACUU--GCCCGGCGGCAGGGGCCACUAAUUGGCAGAACUUCGACGU ------.((.((((((.(((....((((((((.((((.(((((....)))))(((..(.(((((............))--))))))))).))))))))))....))).)))))).))... ( -43.70) >DroSim_CAF1 18598 112 + 1 ------CUCAAAGUUCCGCCCGCCGGUGGUCCACUCCAUUGGCAUCUGCCAAGUCAUGCGGCAAACUUUUAAAGACUU--GCCCGGCGGCAGUGGCCACUAAUUGGCAGAACUUCGACGU ------.((.((((((.(((....(((((.(((((((.(((((....)))))(((..(.(((((............))--))))))))).))))))))))....))).)))))).))... ( -44.10) >DroPer_CAF1 32939 104 + 1 UGUGGGCUCAAAGUUUCAC------GUAGCCC-------UGGCAUUUGCCAAUCCAUGCGGCAAACUUUAAAAGACCCUGUUCCAGUGG---CGGCCACUAAUUGGUGGAAGUUUGACGU ...(((((....(.....)------..)))))-------((((....))))......(((.(((((((.....(.(((((...))).))---)..(((((....)))))))))))).))) ( -29.80) >DroEre_CAF1 29350 112 + 1 ------CUCAAAGUUCCGCCCGCAGGUGGUCCACUCCAUUGGCAUCUGCCAAGUCAUGCGGCAAACUUUUAAAGACUC--GCCCGGCGGCAGUGGCCACUAAUUGGCACAACUUCGACGU ------.((.(((((..(((....(((((.(((((((.(((((....)))))(((..((((...............))--))..))))).))))))))))....)))..))))).))... ( -38.36) >DroYak_CAF1 5690 112 + 1 ------CUCAAAGUUCCGCCCGCAGGUGGUCCACUCCAUUGGCAUCUGCCAAGUCAUGCGGCAAACUUUUAAAGACUC--GCCCGGCGGCAGUGGCCACUAAUUGGCACAACUUCGACGU ------.((.(((((..(((....(((((.(((((((.(((((....)))))(((..((((...............))--))..))))).))))))))))....)))..))))).))... ( -38.36) >consensus ______CUCAAAGUUCCGCCCGCAGGUGGUCCACUCCAUUGGCAUCUGCCAAGUCAUGCGGCAAACUUUUAAAGACUC__GCCCGGCGGCAGUGGCCACUAAUUGGCAGAACUUCGACGU .......((.((((((.(((....(((((.(((((...(((((....)))))(((.((.(((..................)))))..)))))))))))))....))).)))))).))... (-30.23 = -31.37 + 1.14)

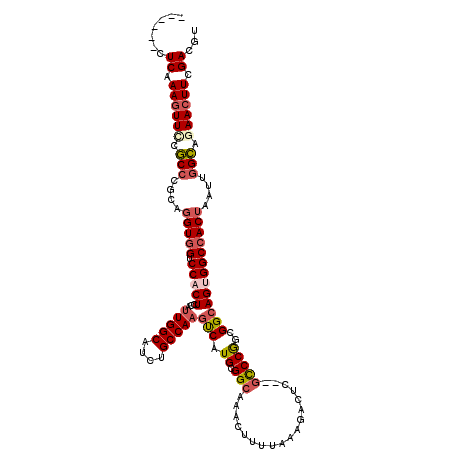
| Location | 1,820,893 – 1,821,005 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -47.78 |
| Consensus MFE | -37.54 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1820893 112 - 22407834 ACGUCGAAGUUCUGCCAAUUAGUGGCCACUGCCGCCGGGC--AAGUCUUUAAAAGUUUGCCGCAUGACUUGGCAGAUGCCAAUGGAGUGGACCACCUGCGGGCGGAACUUUGAG------ ...(((((((((((((.....(((((((((.((((..(((--(((.((.....)))))))))).....(((((....))))).))))))).)))).....))))))))))))).------ ( -53.30) >DroSec_CAF1 40907 112 - 1 ACGUCGAAGUUCUGCCAAUUAGUGGCCCCUGCCGCCGGGC--AAGUCUUUAAAAGUUUGCCGCAUGACUUGGCAGAUGCCAAUGGAGUGGACCACCGGCGGGCGGAACUUUGAG------ ...(((((((((((((.......(((....)))(((((((--(((.((.....)))))))).(((..((((((....))))).)..)))......)))).))))))))))))).------ ( -50.70) >DroSim_CAF1 18598 112 - 1 ACGUCGAAGUUCUGCCAAUUAGUGGCCACUGCCGCCGGGC--AAGUCUUUAAAAGUUUGCCGCAUGACUUGGCAGAUGCCAAUGGAGUGGACCACCGGCGGGCGGAACUUUGAG------ ...(((((((((((((..(..(((((((((.((((..(((--(((.((.....)))))))))).....(((((....))))).))))))).))))..)..))))))))))))).------ ( -54.80) >DroPer_CAF1 32939 104 - 1 ACGUCAAACUUCCACCAAUUAGUGGCCG---CCACUGGAACAGGGUCUUUUAAAGUUUGCCGCAUGGAUUGGCAAAUGCCA-------GGGCUAC------GUGAAACUUUGAGCCCACA ..................(((((((...---)))))))....((((...(((((((((..(((.(((.(((((....))))-------)..))).------))))))))))))))))... ( -30.10) >DroEre_CAF1 29350 112 - 1 ACGUCGAAGUUGUGCCAAUUAGUGGCCACUGCCGCCGGGC--GAGUCUUUAAAAGUUUGCCGCAUGACUUGGCAGAUGCCAAUGGAGUGGACCACCUGCGGGCGGAACUUUGAG------ ...((((((((.((((.....(((((((((.((((..(((--(((.((.....)))))))))).....(((((....))))).))))))).)))).....)))).)))))))).------ ( -48.90) >DroYak_CAF1 5690 112 - 1 ACGUCGAAGUUGUGCCAAUUAGUGGCCACUGCCGCCGGGC--GAGUCUUUAAAAGUUUGCCGCAUGACUUGGCAGAUGCCAAUGGAGUGGACCACCUGCGGGCGGAACUUUGAG------ ...((((((((.((((.....(((((((((.((((..(((--(((.((.....)))))))))).....(((((....))))).))))))).)))).....)))).)))))))).------ ( -48.90) >consensus ACGUCGAAGUUCUGCCAAUUAGUGGCCACUGCCGCCGGGC__AAGUCUUUAAAAGUUUGCCGCAUGACUUGGCAGAUGCCAAUGGAGUGGACCACCUGCGGGCGGAACUUUGAG______ ...((((((((.((((.....(((((((((.(((...(((..............((((((((.......)))))))))))..)))))))).)))).....)))).))))))))....... (-37.54 = -38.30 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:56 2006