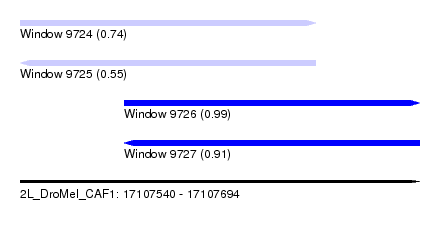
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,107,540 – 17,107,694 |
| Length | 154 |
| Max. P | 0.991278 |

| Location | 17,107,540 – 17,107,654 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -33.79 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107540 114 + 22407834 CGUCUGCAAGACUCCUGAUAAGUGAGCAAACCCAAAGCGAAGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGGACUCACAUACGGCCACUGAGGGAGGGCCAUAC------U .....((....(((((....((((.((.........((....)).........((((((((((((........)))))))......))))).))))))..))))).)).....------. ( -34.20) >DroSec_CAF1 71358 114 + 1 CGUCUGCAAGACUCCUGAUAAGUGAGCAAACCCAAAGCGAAGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGAACUCACAUACGGCCACUGAGGGAGGGUCAUAC------U .((.(((....(((((....((((.((.........((....)).........((((((((((((........)))))))......))))).))))))..))))).).)).))------. ( -33.70) >DroSim_CAF1 84937 114 + 1 CGUCUGCAAGACUCCUGAUAAGUGAGCAAACCCAAAGCGAAGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGAACUCACAUACGGCCACUGAGGGAGGGUCAUAC------U .((.(((....(((((....((((.((.........((....)).........((((((((((((........)))))))......))))).))))))..))))).).)).))------. ( -33.70) >DroEre_CAF1 72761 119 + 1 CGUCUGCAAGACUCCUGAUAAGUGAGCAAACCCAAAGCGAAGGCCAUCACAAAGUAUGCGCUUGGAGAGCCAGCCAAGCGGACUCACAUGCGGCCACU-AUGGAGGGUCAUACUCGCACU .(((.....))).........(((((...((((........((((.......(((...(((((((........))))))).))).......)))).(.-...).))))....)))))... ( -33.24) >DroYak_CAF1 72975 118 + 1 CGUCUGCAAGACUCCUGAUAAGUGAGCAAACCCACAGCGAAGGCCAUCACAAAGUAUGCGCUUGGA--UUCAGCCAAGCGGACUCACAUACGGCAACUGAUGGAGGGUCAUACUCGAACU .(((.....)))..........((((...((((...((....))(((((....((((((((((((.--.....)))))))......)))))(....))))))..))))....)))).... ( -34.10) >consensus CGUCUGCAAGACUCCUGAUAAGUGAGCAAACCCAAAGCGAAGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGGACUCACAUACGGCCACUGAGGGAGGGUCAUAC______U .((.(((....(((((....((((.((.........((....)).........((((((((((((........)))))))......))))).))))))..))))).).)).))....... (-30.46 = -30.90 + 0.44)

| Location | 17,107,540 – 17,107,654 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -33.28 |
| Energy contribution | -33.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107540 114 - 22407834 A------GUAUGGCCCUCCCUCAGUGGCCGUAUGUGAGUCCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCUUCGCUUUGGGUUUGCUCACUUAUCAGGAGUCUUGCAGACG .------...........((..(((((((((....((((.(((((((........)))))))...))))....))))))...)))..))((((((..((((.....))))...)))))). ( -36.40) >DroSec_CAF1 71358 114 - 1 A------GUAUGACCCUCCCUCAGUGGCCGUAUGUGAGUUCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCUUCGCUUUGGGUUUGCUCACUUAUCAGGAGUCUUGCAGACG .------(((.(((((((((..(((((((((....((((.(((((((........)))))))...))))....))))))...)))..)))..............))).))).)))..... ( -37.23) >DroSim_CAF1 84937 114 - 1 A------GUAUGACCCUCCCUCAGUGGCCGUAUGUGAGUUCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCUUCGCUUUGGGUUUGCUCACUUAUCAGGAGUCUUGCAGACG .------(((.(((((((((..(((((((((....((((.(((((((........)))))))...))))....))))))...)))..)))..............))).))).)))..... ( -37.23) >DroEre_CAF1 72761 119 - 1 AGUGCGAGUAUGACCCUCCAU-AGUGGCCGCAUGUGAGUCCGCUUGGCUGGCUCUCCAAGCGCAUACUUUGUGAUGGCCUUCGCUUUGGGUUUGCUCACUUAUCAGGAGUCUUGCAGACG ..((((((((.(((((.....-...(((((((((.((((.(((((((........)))))))...)))))))).)))))........))))))))))((((.....))))...))).... ( -40.19) >DroYak_CAF1 72975 118 - 1 AGUUCGAGUAUGACCCUCCAUCAGUUGCCGUAUGUGAGUCCGCUUGGCUGAA--UCCAAGCGCAUACUUUGUGAUGGCCUUCGCUGUGGGUUUGCUCACUUAUCAGGAGUCUUGCAGACG .....(((((.(((((.....((((.(((((....((((.(((((((.....--.)))))))...))))....)))))....)))).))))))))))...........(((.....))). ( -38.70) >consensus A______GUAUGACCCUCCCUCAGUGGCCGUAUGUGAGUCCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCUUCGCUUUGGGUUUGCUCACUUAUCAGGAGUCUUGCAGACG .......(((.((((((.....(((((((((....((((.(((((((........)))))))...))))....))))))........(((....))))))....))).))).)))..... (-33.28 = -33.52 + 0.24)

| Location | 17,107,580 – 17,107,694 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -29.78 |
| Energy contribution | -29.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
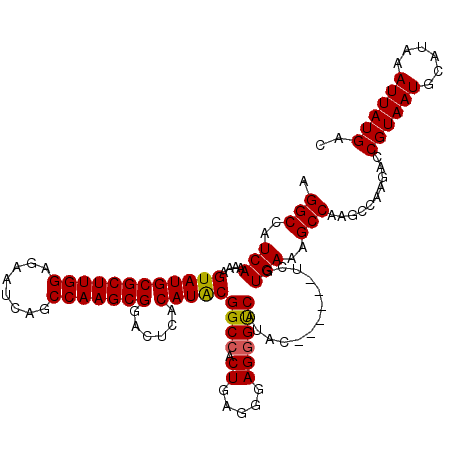
>2L_DroMel_CAF1 17107580 114 + 22407834 AGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGGACUCACAUACGGCCACUGAGGGAGGGCCAUAC------UCGUGAAAGCCAAGCCAAGACCGUAAUGCAUAAAUUAUGAC .(((..(((...(((((.(((((((........)))))))...........((((.((.....))))))))))------)..)))..)))..........((((((......)))))).. ( -34.80) >DroSec_CAF1 71398 114 + 1 AGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGAACUCACAUACGGCCACUGAGGGAGGGUCAUAC------UCGUGAAAGCCAAGCCAAGACCGUAAUGCAUAAAUUAUGAC .(((..(((...(((((.(((((((........)))))))...........((((.((.....))))))))))------)..)))..)))..........((((((......)))))).. ( -31.90) >DroSim_CAF1 84977 114 + 1 AGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGAACUCACAUACGGCCACUGAGGGAGGGUCAUAC------UCGUGAAAGCCAAGCCAAGACCGUAAUGCAUAAAUUAUGAC .(((..(((...(((((.(((((((........)))))))...........((((.((.....))))))))))------)..)))..)))..........((((((......)))))).. ( -31.90) >DroEre_CAF1 72801 119 + 1 AGGCCAUCACAAAGUAUGCGCUUGGAGAGCCAGCCAAGCGGACUCACAUGCGGCCACU-AUGGAGGGUCAUACUCGCACUCGUGAAAGCCAAGUUAAGACCGUAAUGCAUAAAUUAUGAC .(((..((((...((((((((((((........)))))))......)))))((((.((-....))))))............))))..)))..........((((((......)))))).. ( -34.30) >DroYak_CAF1 73015 118 + 1 AGGCCAUCACAAAGUAUGCGCUUGGA--UUCAGCCAAGCGGACUCACAUACGGCAACUGAUGGAGGGUCAUACUCGAACUCGUGAAAGCCAAGCCAAGACCGUAAUGCAUAAAUUAUGAC .(((..((((..(((((.(((((((.--.....)))))))(((((.(((..(....)..)))..)))))))))).(....)))))..)))..........((((((......)))))).. ( -36.50) >consensus AGGCCAUCAAAAAGUAUGCGCUUGGAGAAUCAGCCAAGCGGACUCACAUACGGCCACUGAGGGAGGGUCAUAC______UCGUGAAAGCCAAGCCAAGACCGUAAUGCAUAAAUUAUGAC .(((..(((....((((((((((((........)))))))......)))))((((.((.....)))))).............)))..)))..........((((((......)))))).. (-29.78 = -29.66 + -0.12)

| Location | 17,107,580 – 17,107,694 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.75 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
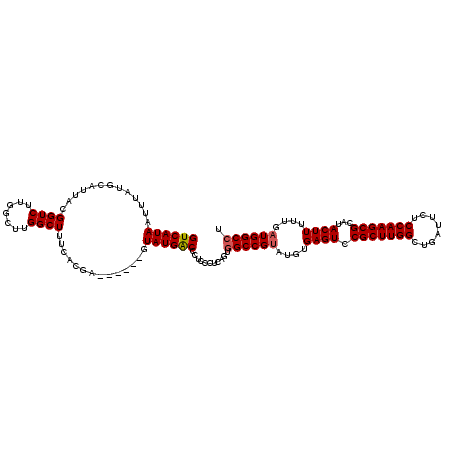
>2L_DroMel_CAF1 17107580 114 - 22407834 GUCAUAAUUUAUGCAUUACGGUCUUGGCUUGGCUUUCACGA------GUAUGGCCCUCCCUCAGUGGCCGUAUGUGAGUCCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCU ((((.((..(((((.....((.(((.(((((.......)))------))((((((((.....)).))))))....))).))((((((........)))))))))))..)).))))..... ( -37.00) >DroSec_CAF1 71398 114 - 1 GUCAUAAUUUAUGCAUUACGGUCUUGGCUUGGCUUUCACGA------GUAUGACCCUCCCUCAGUGGCCGUAUGUGAGUUCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCU ...................((((...(((((.......)))------))..))))..........((((((....((((.(((((((........)))))))...))))....)))))). ( -35.70) >DroSim_CAF1 84977 114 - 1 GUCAUAAUUUAUGCAUUACGGUCUUGGCUUGGCUUUCACGA------GUAUGACCCUCCCUCAGUGGCCGUAUGUGAGUUCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCU ...................((((...(((((.......)))------))..))))..........((((((....((((.(((((((........)))))))...))))....)))))). ( -35.70) >DroEre_CAF1 72801 119 - 1 GUCAUAAUUUAUGCAUUACGGUCUUAACUUGGCUUUCACGAGUGCGAGUAUGACCCUCCAU-AGUGGCCGCAUGUGAGUCCGCUUGGCUGGCUCUCCAAGCGCAUACUUUGUGAUGGCCU (((((((..(((((.....((.((((...((((...(((..(((.(((.......))))))-.)))))))....)))).))((((((........)))))))))))..)))))))..... ( -39.40) >DroYak_CAF1 73015 118 - 1 GUCAUAAUUUAUGCAUUACGGUCUUGGCUUGGCUUUCACGAGUUCGAGUAUGACCCUCCAUCAGUUGCCGUAUGUGAGUCCGCUUGGCUGAA--UCCAAGCGCAUACUUUGUGAUGGCCU (((((((..(((((.....((((...(((((((((....))).))))))..))))(((((((.......).))).)))...((((((.....--.)))))))))))..)))))))..... ( -37.30) >consensus GUCAUAAUUUAUGCAUUACGGUCUUGGCUUGGCUUUCACGA______GUAUGACCCUCCCUCAGUGGCCGUAUGUGAGUCCGCUUGGCUGAUUCUCCAAGCGCAUACUUUUUGAUGGCCU ((((((.............((((.......))))..............))))))...........((((((....((((.(((((((........)))))))...))))....)))))). (-29.51 = -29.75 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:03 2006