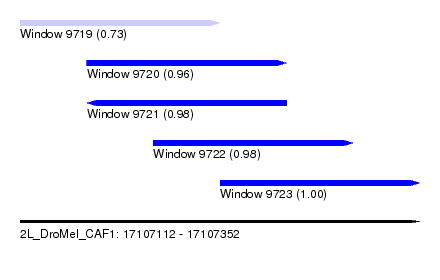
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,107,112 – 17,107,352 |
| Length | 240 |
| Max. P | 0.998605 |

| Location | 17,107,112 – 17,107,232 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -24.79 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107112 120 + 22407834 CAACGGGUGCAGUCUGUAAUUGGAAACUCAUGUUAAUGACUUUCACUCUUUCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCAUAGC ....(((((......(((((.(((((.((((....)))).))))........((((((((..(((....)))..)))))))).......).))))).......)))))............ ( -26.12) >DroSec_CAF1 70930 120 + 1 CAAAGGGUGCAGUCUGUAAUUGGAAACUCAUGUUAAUGACUUUCACUCUCUCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCACAUUACUUCCAUUUACCCCCUUUCCUUAGC ....(((((......(((((((((((.((((....)))).))))........((((((((..(((....)))..))))))))......)).))))).......)))))............ ( -26.22) >DroSim_CAF1 84509 120 + 1 CAAAGGGUGCAGUCUGUAAUUGGAAACUCAUGUUAAUGACUUUCACUCUCUCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCUUAGC .((((((.(......((((.(((((..((((....)))).............((((((((..(((....)))..))))))))..............))))).)))))))))))....... ( -26.20) >DroEre_CAF1 72327 114 + 1 UAAAGGCUGCAGUCUGUAAUUGGCAACUCAUGUUAAUGACUUUCACUC--UCGCCAAAGCCAAAUGAAGGUUAUGCCCUGGCCU----CCCAUUACUUCCAUUUACCCCCAUUCCUUAGC ...(((((((.....))....((((((....))).((((((((((...--..((....))....)))))))))))))..)))))----................................ ( -19.70) >DroYak_CAF1 72543 118 + 1 CAAAGGGUGCAGUCCGUAAUUUGAAACUCAUGUUAAUGACUUACACUC--UCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCACCCAUUACUUCCAUUUACCCCCAUUCCUUAGC ....(((((.((((..((((.((.....)).))))..)))).......--..((((((((..(((....)))..))))))))....)))))............................. ( -25.70) >consensus CAAAGGGUGCAGUCUGUAAUUGGAAACUCAUGUUAAUGACUUUCACUCU_UCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCUUAGC ....(((((......(((((..((((.((((....)))).))))........((((((((..(((....)))..)))))))).........))))).......)))))............ (-21.10 = -22.10 + 1.00)



| Location | 17,107,152 – 17,107,272 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.99 |
| Mean single sequence MFE | -19.39 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107152 120 + 22407834 UUUCACUCUUUCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCAUAGCCUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGA ............((((((((..(((....)))..))))))))..((.((((..................((......))....................((((......)))))))).)) ( -20.50) >DroSec_CAF1 70970 120 + 1 UUUCACUCUCUCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCACAUUACUUCCAUUUACCCCCUUUCCUUAGCCUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGA ............(((...((.....((((((((.....)))))))).)).................................................(((((......))))))))... ( -19.50) >DroSim_CAF1 84549 120 + 1 UUUCACUCUCUCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCUUAGCCUUUCCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGA ............((((((((..(((....)))..))))))))..((.((((..................((......))....................((((......)))))))).)) ( -20.50) >DroEre_CAF1 72367 114 + 1 UUUCACUC--UCGCCAAAGCCAAAUGAAGGUUAUGCCCUGGCCU----CCCAUUACUUCCAUUUACCCCCAUUCCUUAGCCUUACCACUUACAUAUAGCCAUGCAUAUUCAUGGGGCCAG ........--..((...((((.......))))..)).((((((.----.......................................((.......))(((((......))))))))))) ( -17.70) >DroYak_CAF1 72583 118 + 1 UUACACUC--UCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCACCCAUUACUUCCAUUUACCCCCAUUCCUUAGCCUUACCACUUACAUAUACCUAUGCAUAUUCAUGGGGCCAA ........--..((((((((..(((....)))..))))))))........................((((((......((......................))......)))))).... ( -18.75) >consensus UUUCACUCU_UCGCCAAAGCCAAAUGAAGGUUAUGCUUUGGCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCUUAGCCUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGA ............(((..........((((((((.....))))))))....................................................(((((......))))))))... (-17.28 = -17.52 + 0.24)

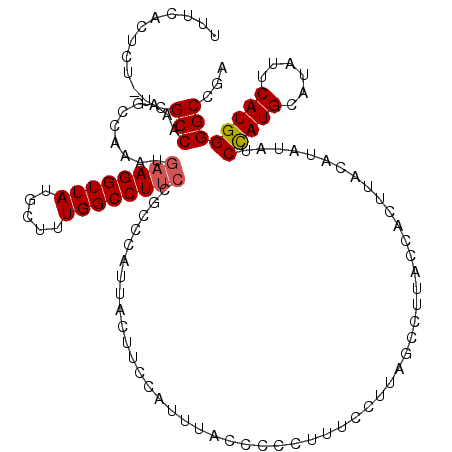
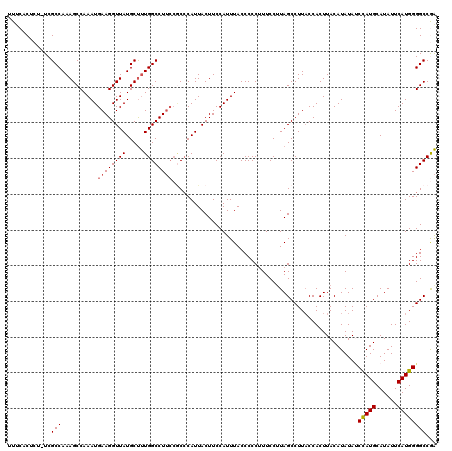
| Location | 17,107,152 – 17,107,272 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.99 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107152 120 - 22407834 UCGGCCCCAUGAAUAUGCAUGGAUAUAUGUAAGUGGUAAGGCUAUGGAAAGGGGGUAAAUGGAAGUAAUGGGCGGAAGGCCAAAGCAUAACCUUCAUUUGGCUUUGGCGAAAGAGUGAAA ...(((((.......((((((....)))))).((((.....))))......)))))........((..(((.(....).)))..))......(((((((.((....))....))))))). ( -29.80) >DroSec_CAF1 70970 120 - 1 UCGGCCCCAUGAAUAUGCAUGGAUAUAUGUAAGUGGUAAGGCUAAGGAAAGGGGGUAAAUGGAAGUAAUGUGCGGAAGGCCAAAGCAUAACCUUCAUUUGGCUUUGGCGAGAGAGUGAAA ((.(((((((.....((((((....)))))).)))).(((((((((..((((..((................(....)((....))))..))))..))))))))))))..))........ ( -28.20) >DroSim_CAF1 84549 120 - 1 UCGGCCCCAUGAAUAUGCAUGGAUAUAUGUAAGUGGAAAGGCUAAGGAAAGGGGGUAAAUGGAAGUAAUGGGCGGAAGGCCAAAGCAUAACCUUCAUUUGGCUUUGGCGAGAGAGUGAAA ((.(((((((.....((((((....)))))).)))).(((((((((..((((..((........((..(((.(....).)))..))))..))))..))))))))))))..))........ ( -32.90) >DroEre_CAF1 72367 114 - 1 CUGGCCCCAUGAAUAUGCAUGGCUAUAUGUAAGUGGUAAGGCUAAGGAAUGGGGGUAAAUGGAAGUAAUGGG----AGGCCAGGGCAUAACCUUCAUUUGGCUUUGGCGA--GAGUGAAA ((.(((((((.....((((((....)))))).)))).((((((...((((((((((...((...((..(((.----...)))..)).))))))))))))))))))))).)--)....... ( -31.50) >DroYak_CAF1 72583 118 - 1 UUGGCCCCAUGAAUAUGCAUAGGUAUAUGUAAGUGGUAAGGCUAAGGAAUGGGGGUAAAUGGAAGUAAUGGGUGGAAGGCCAAAGCAUAACCUUCAUUUGGCUUUGGCGA--GAGUGUAA ((((((((((..((((((....))))))....))))...))))))....(..((.((((((((.((.(((..(((....)))...))).)).)))))))).))..)((..--..)).... ( -32.80) >consensus UCGGCCCCAUGAAUAUGCAUGGAUAUAUGUAAGUGGUAAGGCUAAGGAAAGGGGGUAAAUGGAAGUAAUGGGCGGAAGGCCAAAGCAUAACCUUCAUUUGGCUUUGGCGA_AGAGUGAAA ...(((((((.....((((((....)))))).)))).(((((((((....((((((...((...((..(((.(....).)))..)).)))))))).))))))))))))............ (-26.98 = -27.30 + 0.32)

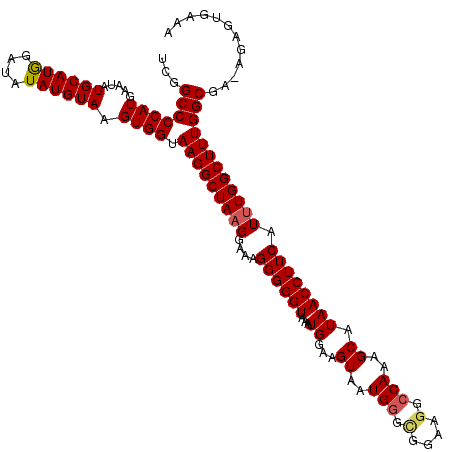

| Location | 17,107,192 – 17,107,312 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107192 120 + 22407834 GCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCAUAGCCUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCC .......(((....(((((........................................((((......))))((((((.........))))))....))))).)))............. ( -22.70) >DroSec_CAF1 71010 120 + 1 GCCUUCCGCACAUUACUUCCAUUUACCCCCUUUCCUUAGCCUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCC (((...........(((((........................................((((......))))((((((.........))))))....))))).)))............. ( -20.70) >DroSim_CAF1 84589 120 + 1 GCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCUUAGCCUUUCCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCC .......(((....(((((........................................((((......))))((((((.........))))))....))))).)))............. ( -22.70) >DroEre_CAF1 72405 116 + 1 GCCU----CCCAUUACUUCCAUUUACCCCCAUUCCUUAGCCUUACCACUUACAUAUAGCCAUGCAUAUUCAUGGGGCCAGCUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCC (((.----......(((((........................................((((......))))(((((...........)))))....))))).)))............. ( -18.90) >consensus GCCUUCCGCCCAUUACUUCCAUUUACCCCCUUUCCUUAGCCUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCC (((...........(((((........................................((((......))))((((((.........))))))....))))).)))............. (-19.58 = -19.82 + 0.25)



| Location | 17,107,232 – 17,107,352 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -20.22 |
| Energy contribution | -19.66 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17107232 120 + 22407834 CUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCCAAUUAGAACUGUAAAAUUAUACUAAUUACAACUUUGAAAU ...................((((......))))((((((.........))))))...(((((((((......).......((((((...((((....)))))))))).)))))))).... ( -22.10) >DroSec_CAF1 71050 120 + 1 CUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCCAAUUAGAACUGUAAAAUUAUACUAAUUACAACUUUGAAAU ...................((((......))))((((((.........))))))...(((((((((......).......((((((...((((....)))))))))).)))))))).... ( -22.10) >DroSim_CAF1 84629 120 + 1 CUUUCCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCCAAUUAGAACUGUAAAAUUAUACUAAUUACAACUUUGAAAU ...................((((......))))((((((.........))))))...(((((((((......).......((((((...((((....)))))))))).)))))))).... ( -22.10) >DroEre_CAF1 72441 120 + 1 CUUACCACUUACAUAUAGCCAUGCAUAUUCAUGGGGCCAGCUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCCAAUUAGAACUGUAAAAUUAUACUAAUUACAACUUUGAAAU .............(((((.((((......))))(((((((((......((.......)).)))))))))...................)))))........................... ( -20.50) >DroYak_CAF1 72661 120 + 1 CUUACCACUUACAUAUACCUAUGCAUAUUCAUGGGGCCAACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCCAAUUAGAACUGUAAAAUUAUACUAAUUACAACUUUGAAAU ...........((((....))))....((((..(((((((((......((.......)).)))))))))...........((((((...((((....)))))))))).......)))).. ( -19.10) >consensus CUUACCACUUACAUAUAUCCAUGCAUAUUCAUGGGGCCGACUCACAUACGGCCCAAACGAAGUUGGCCCAAUCAUCAUCCAAUUAGAACUGUAAAAUUAUACUAAUUACAACUUUGAAAU ........(((((......((((......))))(((((((((......((.......)).)))))))))....................))))).......................... (-20.22 = -19.66 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:59 2006