
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,102,368 – 17,102,624 |
| Length | 256 |
| Max. P | 0.979296 |

| Location | 17,102,368 – 17,102,466 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 98.78 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17102368 98 + 22407834 GGACAGACAGACAGCGUGAUGAACGUCGCCUCAGCUUCGUUACAAUUAACAUAAAUAAUAGCAAAGUGGUCGCUUGCUGUCGCUGUUCUCGUUAUUCC ((((((...((((((((((((((.((.......))))))))))....................((((....)))))))))).)))))).......... ( -28.50) >DroSec_CAF1 66318 98 + 1 GGACAGACAGACAGCGUGAUGAACGUCGCCUCAGCUUCGUUACAAUUAACAUAAAUAAUAGCAAAGUGGUCGCUUGCUGUCACUGUUCUCGUUAUUCC ((((((...((((((((((((((.((.......))))))))))....................((((....)))))))))).)))))).......... ( -26.30) >DroSim_CAF1 62602 98 + 1 GGACAGACAGACAGCGUGAUGAACGUCGCCUCAGCUUCGUUACAAUUAACAUAAAUAAUAGCAAAGUGGUCGCUUGCUGUCGCUGUUCUCGUUAUUCC ((((((...((((((((((((((.((.......))))))))))....................((((....)))))))))).)))))).......... ( -28.50) >DroEre_CAF1 67523 98 + 1 GGACAGACAGACAGCGUGAUGAACGUCGCCUCAGCUUCGUUACAAUUAACAUAAAUAAUAGCAAAGUGGUCGCUUGCUGUCGCUGUUCUCGUUAUUCC ((((((...((((((((((((((.((.......))))))))))....................((((....)))))))))).)))))).......... ( -28.50) >DroYak_CAF1 66781 98 + 1 AGACAGACAGACAGCGUGAUGAACGUCGCCUCAGCUUCGUUACAAUUAACAUAAAUAAUAGCAAAGUGGUCGCUUGCUGUCGCUCUUCUCGUUAUUCC (((.(((..((((((((((((((.((.......))))))))))....................((((....))))))))))..))))))......... ( -22.60) >consensus GGACAGACAGACAGCGUGAUGAACGUCGCCUCAGCUUCGUUACAAUUAACAUAAAUAAUAGCAAAGUGGUCGCUUGCUGUCGCUGUUCUCGUUAUUCC ((((((...((((((((((((((.((.......))))))))))....................((((....)))))))))).)))))).......... (-26.16 = -26.56 + 0.40)

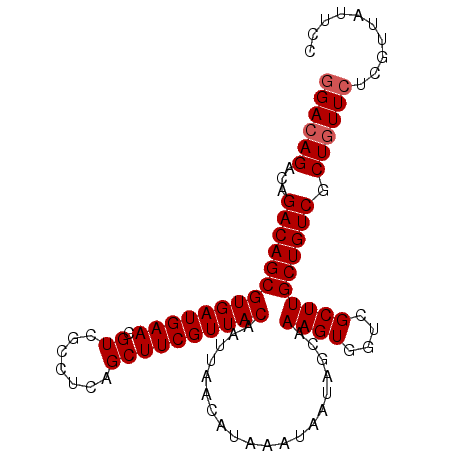
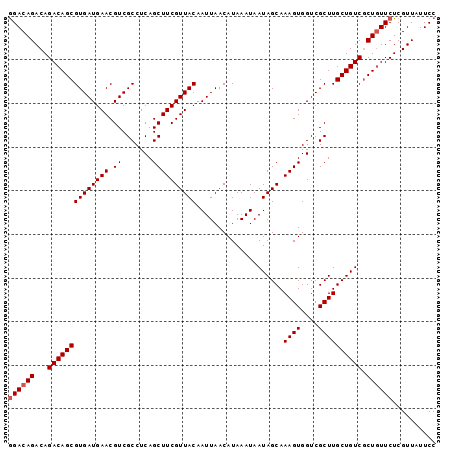
| Location | 17,102,466 – 17,102,569 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -24.25 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17102466 103 + 22407834 -----------------CUUUGUUGCUGCUGAAAAGUGGAAAUAUAUUUUUCACUUUUCAUUUCUAAUUAUGACGCCGGGGACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCA -----------------..((((.((...(((((((((((((.....))))))))))))).................((((.....))))....)).))))(((((....)))))..... ( -27.40) >DroPse_CAF1 58703 120 + 1 CUGGCUCUCGUUGUUGUCGUUGUUUUUGCUGAAAAGUGGAAAUAUAUUUUUCACUUUUCAUUUCUAAUUAUGACGCCGGGGACAUGCUCCAAAUGCCACAAACAAAAUGUUUUGUAUUUU .((((.......(.((((((.(((.....(((((((((((((.....))))))))))))).....))).)))))).)((((.....))))....))))...(((((....)))))..... ( -31.90) >DroSec_CAF1 66416 103 + 1 -----------------CUUUGUUGCUGCUGAAAAGUGGAAAUAUAUUUUUCACUUUUCAUUUCUAAUUAUGACGCCGGGGACAUGCCCCAAAUGCCAAAAACAAAAUGUUUUGUAUUCA -----------------.((((((...(((((((((((((((.....))))))))))))).................((((.....))))....))....)))))).............. ( -26.60) >DroEre_CAF1 67621 103 + 1 -----------------CUUUGUUGCUGCUGAAAAGUGGAAAUAUAUUUUUCACUUUUCAUUUCUAAUUAUGACGCCGGGGACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCA -----------------..((((.((...(((((((((((((.....))))))))))))).................((((.....))))....)).))))(((((....)))))..... ( -27.40) >DroYak_CAF1 66879 101 + 1 -----------------CUUUGUAGCUGCUGAAAAGUGGAAAU--AUUUUUCACUUUUCAUUUCUAAUUAUGACGCUGGGGACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCA -----------------..((((.((.((((((((((((((..--...))))))))))))..((.......)).))(((((.....)))))...)).))))(((((....)))))..... ( -26.40) >DroPer_CAF1 57042 120 + 1 CUGGCUCUCGUUGUUGUCGUUGUUUUUGCUGAAAAGUGGAAAUAUAUUUUUCACUUUUCAUUUCUAAUUAUGACGCCGGGGACAUGCUCCAAAUGCCACAAACAAAAUGUUUUGUAUUUU .((((.......(.((((((.(((.....(((((((((((((.....))))))))))))).....))).)))))).)((((.....))))....))))...(((((....)))))..... ( -31.90) >consensus _________________CUUUGUUGCUGCUGAAAAGUGGAAAUAUAUUUUUCACUUUUCAUUUCUAAUUAUGACGCCGGGGACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCA ...................(((((...(((((((((((((((.....))))))))))))).................((((.....))))....))....)))))............... (-24.25 = -24.20 + -0.05)

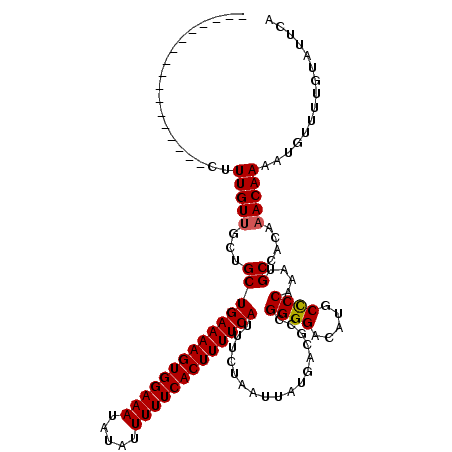

| Location | 17,102,466 – 17,102,569 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17102466 103 - 22407834 UGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUUUCCACUUUUCAGCAGCAACAAAG----------------- .............(((((.(((((((..((((((.....)))))).))))))).))))).(((((((((.(((.....))).)))))))))............----------------- ( -25.40) >DroPse_CAF1 58703 120 - 1 AAAAUACAAAACAUUUUGUUUGUGGCAUUUGGAGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUUUCCACUUUUCAGCAAAAACAACGACAACAACGAGAGCCAG ........((((.....)))).((((.((((..((.((....))))(((...........(((((((((.(((.....))).)))))))))...........))).....)))).)))). ( -22.45) >DroSec_CAF1 66416 103 - 1 UGAAUACAAAACAUUUUGUUUUUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUUUCCACUUUUCAGCAGCAACAAAG----------------- ..............((((((.(((((....((((.....)))).))..............(((((((((.(((.....))).))))))))).))).)))))).----------------- ( -22.70) >DroEre_CAF1 67621 103 - 1 UGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUUUCCACUUUUCAGCAGCAACAAAG----------------- .............(((((.(((((((..((((((.....)))))).))))))).))))).(((((((((.(((.....))).)))))))))............----------------- ( -25.40) >DroYak_CAF1 66879 101 - 1 UGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCAGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAU--AUUUCCACUUUUCAGCAGCUACAAAG----------------- ..................((((((((...(((((.....)))))((.((.......))..((((((((((((...--.))).))))))))))).)))))))).----------------- ( -28.20) >DroPer_CAF1 57042 120 - 1 AAAAUACAAAACAUUUUGUUUGUGGCAUUUGGAGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUUUCCACUUUUCAGCAAAAACAACGACAACAACGAGAGCCAG ........((((.....)))).((((.((((..((.((....))))(((...........(((((((((.(((.....))).)))))))))...........))).....)))).)))). ( -22.45) >consensus UGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUUUCCACUUUUCAGCAGCAACAAAG_________________ .............(((((.(((((((..((((((.....)))))).))))))).))))).(((((((((.(((.....))).)))))))))............................. (-22.57 = -22.93 + 0.36)

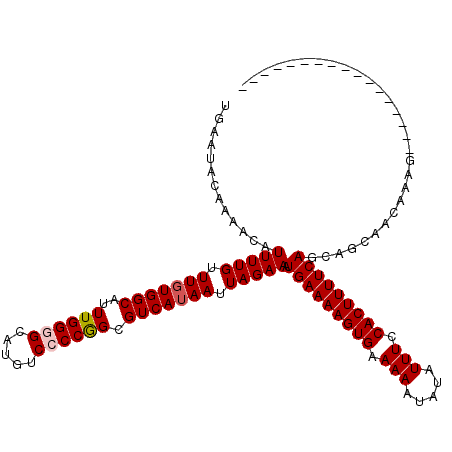
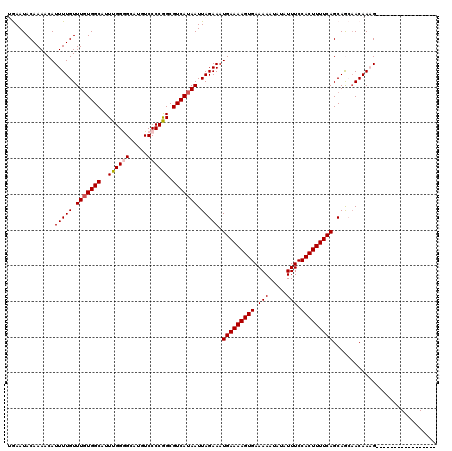
| Location | 17,102,489 – 17,102,585 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17102489 96 - 22407834 CCGCACUACAAAAU------------------------GCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUU .(((......((((------------------------((((...(((((....)))))...))))))))((((.....)))).)))((((..(((....)))...)))).......... ( -22.70) >DroPse_CAF1 58743 120 - 1 CCGCAUCACAAUGUGCAGAAGCAGAAGCAAAAGCAAUAGCAAAAUACAAAACAUUUUGUUUGUGGCAUUUGGAGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUU .....((((..((((.....((....))....((....))....))))...((((((..(((((((.(..((........))..).)))))))...))))))....)))).......... ( -21.80) >DroSec_CAF1 66439 96 - 1 CCGCACAACAAAAU------------------------GCUGAAUACAAAACAUUUUGUUUUUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUU .(((......((((------------------------((..((.(((((....))))).))..))))))((((.....)))).)))((((..(((....)))...)))).......... ( -25.40) >DroEre_CAF1 67644 96 - 1 CCGCACAACAAAAU------------------------GCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUU .(((......((((------------------------((((...(((((....)))))...))))))))((((.....)))).)))((((..(((....)))...)))).......... ( -22.70) >DroYak_CAF1 66902 94 - 1 CCGCACAACAAAAU------------------------GCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCAGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAU--AUU .(((......((((------------------------((((...(((((....)))))...))))))))((((.....)))).)))((((..(((....)))...)))).....--... ( -22.90) >DroPer_CAF1 57082 120 - 1 CCGCAUCACAAUGUGCAGAAGCAGAAGCAAAAGCAAUAGCAAAAUACAAAACAUUUUGUUUGUGGCAUUUGGAGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUU .....((((..((((.....((....))....((....))....))))...((((((..(((((((.(..((........))..).)))))))...))))))....)))).......... ( -21.80) >consensus CCGCACAACAAAAU________________________GCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUCCCCGGCGUCAUAAUUAGAAAUGAAAAGUGAAAAAUAUAUU ...................................................((((((..(((((((..((((((.....)))))).)))))))...)))))).................. (-16.27 = -16.63 + 0.36)

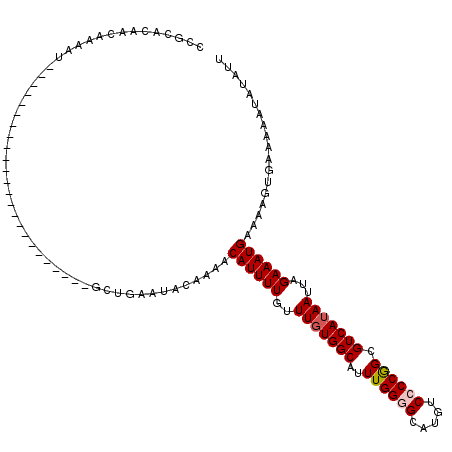
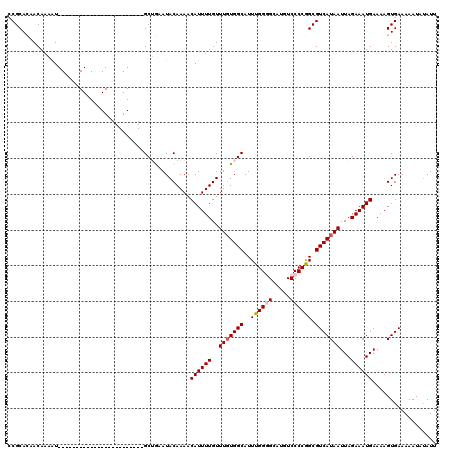
| Location | 17,102,529 – 17,102,624 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.52 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -20.02 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17102529 95 + 22407834 GACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCAGCAUUUUGUAGUGCGGCCACAGAGUGAAAGGUGCGUUAAAUUUGUUAAAUGGAUA ...((((.((....(((.((.((((((((((........))))))))))..)).)))(((...)))...)).))))................... ( -20.90) >DroSec_CAF1 66479 95 + 1 GACAUGCCCCAAAUGCCAAAAACAAAAUGUUUUGUAUUCAGCAUUUUGUUGUGCGGCCACAGAGUGAAAGGUGCGUAAAAUUUGUAAAAUGGAUA ...((((.((....(((...(((((((((((........)))))))))))....)))(((...)))...)).))))................... ( -20.70) >DroSim_CAF1 62763 95 + 1 GACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCAGCAUUUUGUUGUGCGGCCACAGAGUGAAAGGUGCGUUAAAUUUGUAAAAUGGAUA ...((((.((....(((.(((((((((((((........))))))))))).)).)))(((...)))...)).))))................... ( -22.00) >DroEre_CAF1 67684 94 + 1 GACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCAGCAUUUUGUUGUGCGGCCACAGAGUGAAAGGUGCGUUACAUUU-UAUAAUUGAUA ...((((.((....(((.(((((((((((((........))))))))))).)).)))(((...)))...)).)))).......-........... ( -22.00) >DroYak_CAF1 66940 94 + 1 GACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCAGCAUUUUGUUGUGCGGUCACAGAGUGAAAGGUGUGUUACAUUU-CAAAAUGGACA ((((((((.....(((.((((.(((((((((........)))))))))))))))).((((...))))..)))))))).((((.-...)))).... ( -24.10) >consensus GACAUGCCCCAAAUGCCACAAACAAAAUGUUUUGUAUUCAGCAUUUUGUUGUGCGGCCACAGAGUGAAAGGUGCGUUAAAUUUGUAAAAUGGAUA ...((((.((....(((.(((((((((((((........))))))))))).)).)))(((...)))...)).))))................... (-20.02 = -20.10 + 0.08)

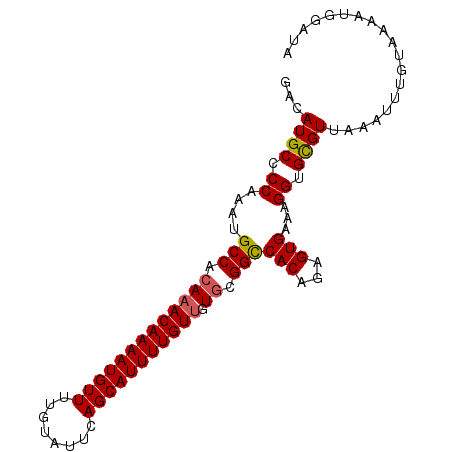
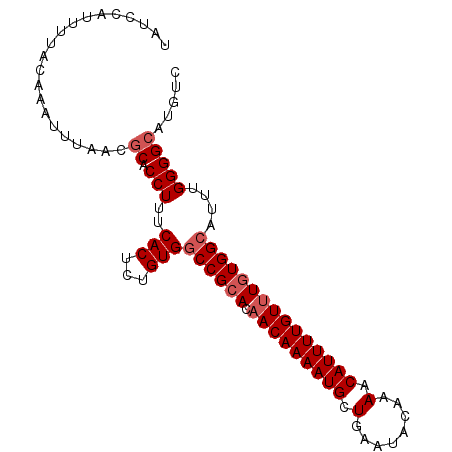
| Location | 17,102,529 – 17,102,624 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.52 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -18.38 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17102529 95 - 22407834 UAUCCAUUUAACAAAUUUAACGCACCUUUCACUCUGUGGCCGCACUACAAAAUGCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUC .....................((.(((..(((...)))((((((..((((((((.(........).)))))))).))))))....)))))..... ( -22.00) >DroSec_CAF1 66479 95 - 1 UAUCCAUUUUACAAAUUUUACGCACCUUUCACUCUGUGGCCGCACAACAAAAUGCUGAAUACAAAACAUUUUGUUUUUGGCAUUUGGGGCAUGUC .....................((.(((.......((((....))))...((((((..((.(((((....))))).))..)))))))))))..... ( -20.70) >DroSim_CAF1 62763 95 - 1 UAUCCAUUUUACAAAUUUAACGCACCUUUCACUCUGUGGCCGCACAACAAAAUGCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUC .....................((.(((..(((...)))((((((.(((((((((.(........).)))))))))))))))....)))))..... ( -22.80) >DroEre_CAF1 67684 94 - 1 UAUCAAUUAUA-AAAUGUAACGCACCUUUCACUCUGUGGCCGCACAACAAAAUGCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUC ...........-.........((.(((..(((...)))((((((.(((((((((.(........).)))))))))))))))....)))))..... ( -22.80) >DroYak_CAF1 66940 94 - 1 UGUCCAUUUUG-AAAUGUAACACACCUUUCACUCUGUGACCGCACAACAAAAUGCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUC (((((....((-(((.((.....)).)))))(...(((.(((((.(((((((((.(........).)))))))))))))))))..)))))).... ( -24.40) >consensus UAUCCAUUUUACAAAUUUAACGCACCUUUCACUCUGUGGCCGCACAACAAAAUGCUGAAUACAAAACAUUUUGUUUGUGGCAUUUGGGGCAUGUC .....................((.(((..(((...)))((((((.(((((((((.(........).)))))))))))))))....)))))..... (-18.38 = -19.18 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:34 2006