
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,101,425 – 17,101,569 |
| Length | 144 |
| Max. P | 0.973841 |

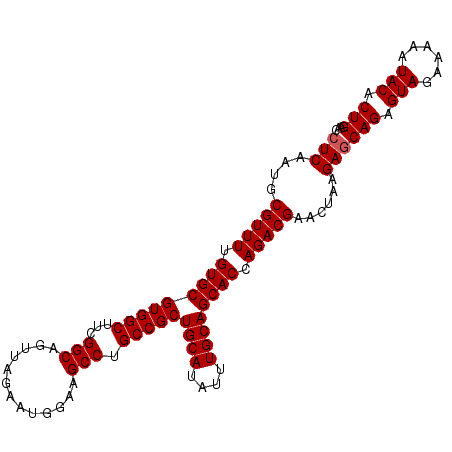
| Location | 17,101,425 – 17,101,529 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -31.09 |
| Energy contribution | -31.34 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17101425 104 + 22407834 CUCAAUGCGUUUUGUGCGUGGCUUCGGCAGUGAGACUGGAAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUAAGAGCAGAGUAGAAAAAUACACUGAAC (((....(((((.(((((.((((((..(((.....)))))))))))).((((((....)))))))).)))))......)))(((.(((......))).)))... ( -34.60) >DroSec_CAF1 65413 104 + 1 CUCAAUGCGUUUUGUGCGUGGCUUCGGCAGUUAGACUGGAAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUAAGAGCAGAGUAGAAAAAUACACUGAGC (((....(((((.(((((.((((((..(((.....)))))))))))).((((((....)))))))).)))))......)))(((.(((......))).)))... ( -34.60) >DroEre_CAF1 66568 101 + 1 CUCAAUGCGUUUUGUGCGUGGCUUUGGCAGUUAGAAUGGUAGCCUGCCGCUGCAUAUUUGCAGCAC-AGACGCACUAAGAGCAGAGUAGA-AAGUACACUGAA- (((..(((((((.(((((((((...(((..((.....))..))).)))))((((....))))))))-)))))))....)))(((.(((..-...))).)))..- ( -35.50) >DroYak_CAF1 65774 104 + 1 CUCAAUGCGUUUUGUGCGUGGCUUUGGCAGUUAGAAUGGGAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUACGACCAGAGUAGAGAAAUACACUGAAC .(((.(((((((.(((((((((...(((.............))).)))))((((....)))))))).)))))..((((.......)))).......)).))).. ( -32.52) >consensus CUCAAUGCGUUUUGUGCGUGGCUUCGGCAGUUAGAAUGGAAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUAAGAGCAGAGUAGAAAAAUACACUGAAC (((....(((((.(((((((((...(((.............))).)))))((((....)))))))).)))))......)))(((.(((......))).)))... (-31.09 = -31.34 + 0.25)

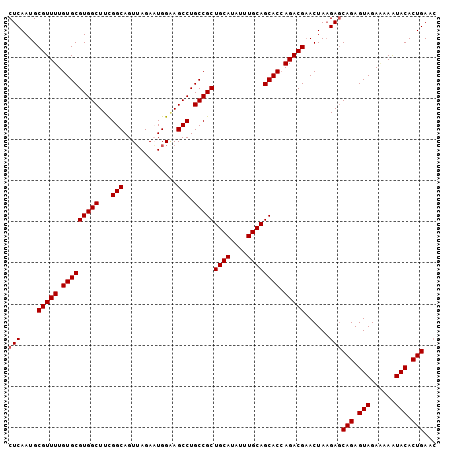
| Location | 17,101,425 – 17,101,529 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -29.61 |
| Energy contribution | -29.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17101425 104 - 22407834 GUUCAGUGUAUUUUUCUACUCUGCUCUUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUUCCAGUCUCACUGCCGAAGCCACGCACAAAACGCAUUGAG ..((((((..((((.......(((............((.(((((((....))))))).))(((((((((.....)))..))))))..))).))))..)))))). ( -33.20) >DroSec_CAF1 65413 104 - 1 GCUCAGUGUAUUUUUCUACUCUGCUCUUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUUCCAGUCUAACUGCCGAAGCCACGCACAAAACGCAUUGAG .(((((((..((((.......(((............((.(((((((....))))))).))(((((((((.....)))..))))))..))).))))..))))))) ( -35.00) >DroEre_CAF1 66568 101 - 1 -UUCAGUGUACUU-UCUACUCUGCUCUUAGUGCGUCU-GUGCUGCAAAUAUGCAGCGGCAGGCUACCAUUCUAACUGCCAAAGCCACGCACAAAACGCAUUGAG -..(((.(((...-..))).)))..((((((((((.(-((((((((....))))))(((((.............)))))..........)))..)))))))))) ( -34.12) >DroYak_CAF1 65774 104 - 1 GUUCAGUGUAUUUCUCUACUCUGGUCGUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUCCCAUUCUAACUGCCAAAGCCACGCACAAAACGCAUUGAG ..(((((((......((((.......))))...((.((((((((((....))))))(((((.............)))))...)))).)).......))))))). ( -33.92) >consensus GUUCAGUGUAUUUUUCUACUCUGCUCUUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUUCCAGUCUAACUGCCAAAGCCACGCACAAAACGCAUUGAG .((((((((........(((........)))..((.((((((((((....))))))(((((.............)))))...)))).)).......)))))))) (-29.61 = -29.67 + 0.06)


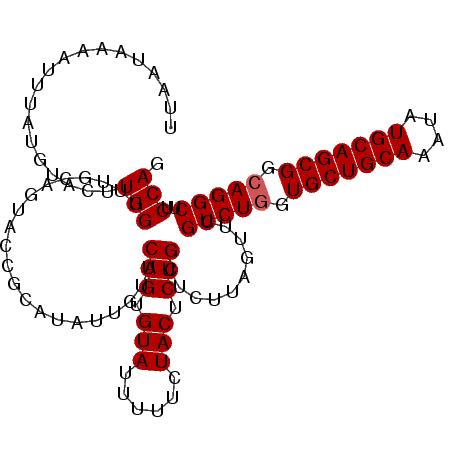
| Location | 17,101,460 – 17,101,569 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17101460 109 + 22407834 CUGGAAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUAAGAGCAGAGUAGAAAAAUACACUGAACAAAAUGCGGUUCUGCAAACCAAGUACAUAAAUUUUAUUAA .(((..(.(((..((((((....))))))..))).).........((((((((......)))((((.........)))))))))...)))................... ( -25.00) >DroSec_CAF1 65448 109 + 1 CUGGAAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUAAGAGCAGAGUAGAAAAAUACACUGAGCAAUAUGCAGUACUGCAAACCAAGUACAUAAAUUUUAUUAA ......(((((..((((((....))))))..))).........(..(((.(((......))).)))..).....)).(((((........))))).............. ( -25.90) >DroEre_CAF1 66603 95 + 1 AUGGUAGCCUGCCGCUGCAUAUUUGCAGCAC-AGACGCACUAAGAGCAGAGUAGA-AAGUACACUGAA------------CUGCAAACCAUGCAGAUACUUUUUAUUAA .((((.(((((..((((((....)))))).)-))..)))))).......((((((-(((((.......------------(((((.....))))).))))))))))).. ( -27.50) >DroYak_CAF1 65809 109 + 1 AUGGGAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUACGACCAGAGUAGAGAAAUACACUGAACAAUUCGUGGUACUACAAGCCAAGUACAUAAAUUUUAUUAA .(((..(.(((..((((((....))))))..))).)..(((((((.(((.(((......))).)))......)))))))........)))................... ( -26.10) >consensus AUGGAAGCCUGCCGCUGCAUAUUUGCAGCACCAGACGAACUAAGAGCAGAGUAGAAAAAUACACUGAACAAUAUGCGGUACUGCAAACCAAGUACAUAAAUUUUAUUAA .(((..(.(((..((((((....))))))..))).)..........(((.(((......))).))).....................)))................... (-18.35 = -18.60 + 0.25)



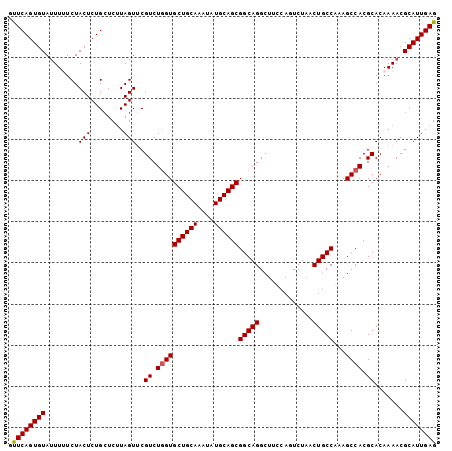
| Location | 17,101,460 – 17,101,569 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17101460 109 - 22407834 UUAAUAAAAUUUAUGUACUUGGUUUGCAGAACCGCAUUUUGUUCAGUGUAUUUUUCUACUCUGCUCUUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUUCCAG ....................(((((...)))))(.(((..(..(((.(((......))).)))..)..))).)(((((.(((((((....))))))).)))))...... ( -28.30) >DroSec_CAF1 65448 109 - 1 UUAAUAAAAUUUAUGUACUUGGUUUGCAGUACUGCAUAUUGCUCAGUGUAUUUUUCUACUCUGCUCUUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUUCCAG ...................(((...((((((.....)))))).(((.(((......))).)))..........(((((.(((((((....))))))).)))))..))). ( -29.40) >DroEre_CAF1 66603 95 - 1 UUAAUAAAAAGUAUCUGCAUGGUUUGCAG------------UUCAGUGUACUU-UCUACUCUGCUCUUAGUGCGUCU-GUGCUGCAAAUAUGCAGCGGCAGGCUACCAU ..........((..(((((((.(((((((------------(.(((((((((.-..............)))))).))-).)))))))))))))))..)).......... ( -30.76) >DroYak_CAF1 65809 109 - 1 UUAAUAAAAUUUAUGUACUUGGCUUGUAGUACCACGAAUUGUUCAGUGUAUUUCUCUACUCUGGUCGUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUCCCAU ..............(((((........)))))...((((((.((((.(((......))).))))...))))))(((((.(((((((....))))))).)))))...... ( -33.00) >consensus UUAAUAAAAUUUAUGUACUUGGUUUGCAGUACCGCAUAUUGUUCAGUGUAUUUUUCUACUCUGCUCUUAGUUCGUCUGGUGCUGCAAAUAUGCAGCGGCAGGCUUCCAG ...................(((.....................(((.(((......))).)))..........(((((.(((((((....))))))).)))))..))). (-24.08 = -24.32 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:25 2006