
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
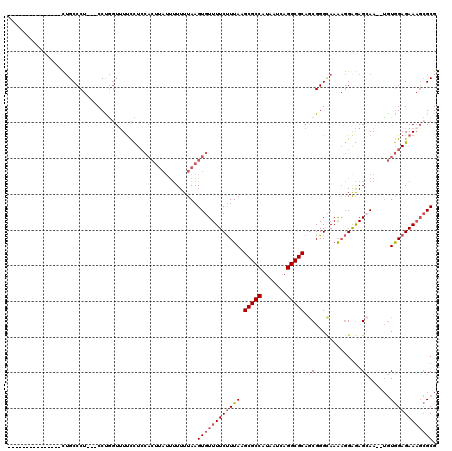
| Location | 17,096,615 – 17,096,749 |
| Length | 134 |
| Max. P | 0.768973 |

| Location | 17,096,615 – 17,096,715 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -20.34 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17096615 100 + 22407834 ---------------CUGCCCU---CCUGCUUUUCCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAA--UGUGGAGAAUGCGCG ---------------.(((.((---(((..((.(((..((((((......))))))..........(((((.......)))))...))).)).))))).))).--.(((.......))). ( -30.60) >DroPse_CAF1 53017 113 + 1 CGGUAUAUCGACUGAUAGCCCUCUUCCUUGUUUCUUUC-------CGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACGGGAAAAACGAAAGAAAGAGAUAGAGAAAUAGCG .(((.((((....)))))))((((..(((.((((((((-------.(((((........((((...(((((.......)))))...))))))))))))))))))))..))))........ ( -32.70) >DroSec_CAF1 60292 100 + 1 ---------------CUGCCCU---CCUGGUUUUCCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAA--UGUGGAGAAAGCGCG ---------------..((.((---((..((((((((.((((((......))))))..........(((((.......)))))..........))))))))..--...))))...))... ( -31.60) >DroSim_CAF1 56589 100 + 1 ---------------CUGCCCU---CCUAGUUUUCCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAA--UGUGGAGAAAGCGCG ---------------.......---....((((((.((((((((......))).((((((((((..(((((.......))))).(....)..)))))))))).--.)))))))))))... ( -30.60) >DroEre_CAF1 61682 100 + 1 ---------------CUGCCCU---CCCGGUCUUCCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGUAAAAGGAGAGCAA--UGUGGAGAAAGCGCG ---------------.(((.((---((...(..(((..((((((......))))))..........(((((.......)))))...)))..)..)))).))).--.(((.......))). ( -28.30) >DroYak_CAF1 60777 100 + 1 ---------------CUGCCCU---CCCGGUUUUCCUGCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGUAAGCGGGGAGGAA--UGUGGAGAAAGCGCG ---------------....(((---(((.((((.((((((((((......)))))...........(((((.......))))).))))).)))).))))))..--.(((.......))). ( -41.40) >consensus _______________CUGCCCU___CCUGGUUUUCCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAA__UGUGGAGAAAGCGCG .....................................................((((((((((((.(((((.......))))).((.............))......)))))))))))). (-20.34 = -21.20 + 0.86)


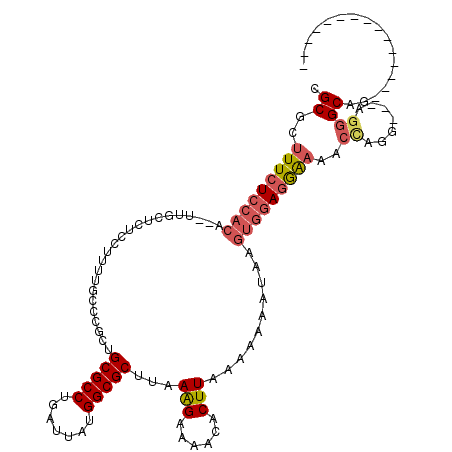
| Location | 17,096,615 – 17,096,715 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -19.27 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17096615 100 - 22407834 CGCGCAUUCUCCACA--UUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGGAAAAGCAGG---AGGGCAG--------------- ...............--.((((((((((((..((...(((((.......)))))..........((((((......))))))..))..))).)))---)))))).--------------- ( -33.40) >DroPse_CAF1 53017 113 - 1 CGCUAUUUCUCUAUCUCUUUCUUUCGUUUUUCCCGUUGCGCCUGAUUAUGGCGCUUAAGGAAAACACUUAAAACG-------GAAAGAAACAAGGAAGAGGGCUAUCAGUCGAUAUACCG ((.(((.((.....(((((((((...(((((.((((((((((.......)))))(((((.......)))))))))-------).)))))..)))))))))(((.....))))).))).)) ( -34.60) >DroSec_CAF1 60292 100 - 1 CGCGCUUUCUCCACA--UUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGGAAAACCAGG---AGGGCAG--------------- ...............--.((((((((((((..((...(((((.......)))))..........((((((......))))))..)).)))..)))---)))))).--------------- ( -31.80) >DroSim_CAF1 56589 100 - 1 CGCGCUUUCUCCACA--UUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGGAAAACUAGG---AGGGCAG--------------- ...............--.(((((((((.((..((...(((((.......)))))..........((((((......))))))..))..))..)))---)))))).--------------- ( -32.50) >DroEre_CAF1 61682 100 - 1 CGCGCUUUCUCCACA--UUGCUCUCCUUUUACCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGGAAGACCGGG---AGGGCAG--------------- ...............--.((((((((((((.(((...(((((.......)))))..........((((((......))))))).)).)))..)))---)))))).--------------- ( -31.50) >DroYak_CAF1 60777 100 - 1 CGCGCUUUCUCCACA--UUCCUCCCCGCUUACCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGCAGGAAAACCGGG---AGGGCAG--------------- ...((((((((....--(((((....((......)).(((((.......)))))..........((((((......)))))).)))))....)))---)))))..--------------- ( -30.30) >consensus CGCGCUUUCUCCACA__UUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGGAAAACCAGG___AGGGCAG_______________ .((..(((((((((.......................(((((.......)))))...(((......)))..........)))))))))..((.......))))................. (-19.27 = -20.08 + 0.81)

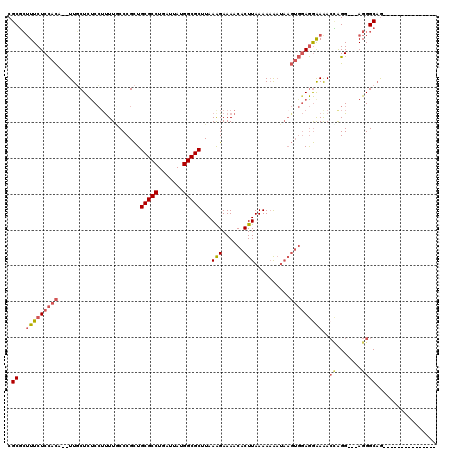
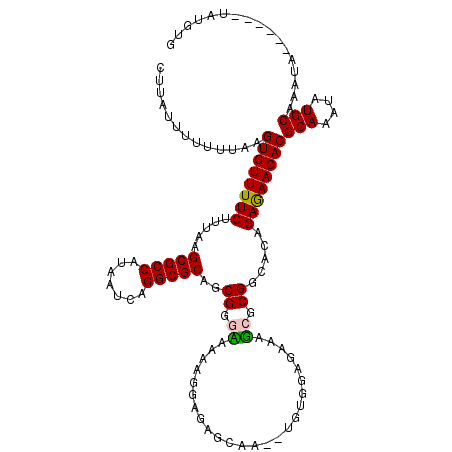
| Location | 17,096,637 – 17,096,749 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.46 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17096637 112 + 22407834 CUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAA--UGUGGAGAAUGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUAUG .............(((((((((.((.(((((.......)))))..((.(((............--.........))).)).)).)))))))))(((....))).....------...... ( -27.50) >DroPse_CAF1 53055 109 + 1 -----CGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACGGGAAAAACGAAAGAAAGAGAUAGAGAAAUAGCGGCAUAGAAAACACGCAAAUAUGCAAAUG------UAUGUG -----.(((((..((((((((((...(((((.......)))))...))))))..(....)....................))))..))))).((((.((((....)))------).)))) ( -27.40) >DroSim_CAF1 56611 112 + 1 CUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAA--UGUGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUGUG ....(((((.((.((((((((((((.(((((.......))))).((...(.....)...))..--..))))))))))))..)).)))))...((((.((((....)))------).)))) ( -30.20) >DroEre_CAF1 61704 118 + 1 CUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGUAAAAGGAGAGCAA--UGUGGAGAAAGCGCGGCACGGAGAACACGCAAAUAUGCAAAUACACUUGUAUGUG ....((((((((..((((((((((..(((((.......))))).(....)..)))))))))).--..))))))))(((.(..........).)))..((((((((......)))))))). ( -30.60) >DroYak_CAF1 60799 112 + 1 CUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGUAAGCGGGGAGGAA--UGUGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUGUG ....(((((.((.((((((((((((.(((((.......))))).((......)).........--..))))))))))))..)).)))))...((((.((((....)))------).)))) ( -31.00) >DroPer_CAF1 51475 109 + 1 -----CGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACGAGAAAAACGAAAGAAAGAGAUAGAGAAAUAGCGGCAUAGAAAACACGCAAAUAUGCAAAUG------UAUGUG -----........((((((((.....(((((.......)))))...........(....).........................))))))))....((((((....)------))))). ( -24.80) >consensus CUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGAAAAAGGAGAGCAA__UGUGGAGAAAGCGCGGCACAGAGAACACGCAAAUAUGCAAAUA______UAUGUG .............((((((((.....(((((.......)))))..((.((.........................)).)).....))))))))(((....)))................. (-22.90 = -22.46 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:15 2006