
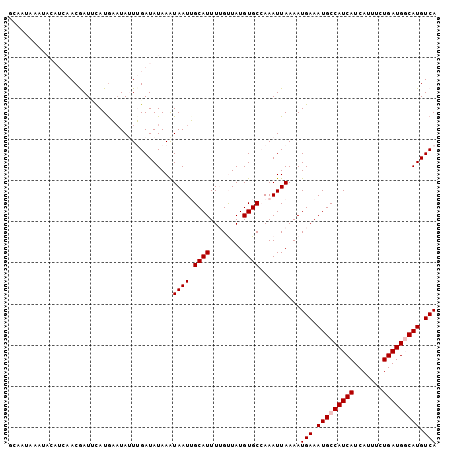
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,089,987 – 17,090,107 |
| Length | 120 |
| Max. P | 0.884811 |

| Location | 17,089,987 – 17,090,091 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17089987 104 + 22407834 GCAAUAAAUACAUCAACAAUUCAUGAUUAUUUGAUAUAAAUAAUUGCAUUUUGUUAUGUGCCAAAUUAAAAUGAAAUGCCAUCAUCAUUUCUGAUGGCAUGUCA ........(((((.(((((.....(((((((((...))))))))).....))))))))))...........(((.(((((((((.......))))))))).))) ( -24.00) >DroPse_CAF1 44787 88 + 1 ---------------ACGAU-UAUGUAUAGUGGAUAUAAAUAAUGGCAUUUGGCUAUGUGCAAAAUUAAAAUGAAAUGCCAUCAUCAUUUUUGAUGCCAUGUCA ---------------.....-........((((.........(((((((((.((.....))............)))))))))(((((....))))))))).... ( -16.92) >DroSec_CAF1 53731 104 + 1 GCAAUAAAUACAUCAACGAUUCAUAAAUAUUUGAUAUAAAUAAUUGCAUUUUGUUAUGUGCCAAAUUAAAAUGAAAUGCCAUCAUCAUUUCUGAUGGCAUGUCA ........(((((.(((((....(((.((((((...)))))).)))....))))))))))...........(((.(((((((((.......))))))))).))) ( -20.80) >DroEre_CAF1 55118 104 + 1 GCAAUAAAUACAUCAACAAUCCAUGAAUAUUUGAUAUAAAUAAUUGCAUUUUGUUAUGUGCCAAAUUAAAAUGAAAUGCCAUCAUCAUUUCUGAUGGCAUGUCA ........(((((.(((((...(((..((((((...))))))....))).))))))))))...........(((.(((((((((.......))))))))).))) ( -21.10) >DroYak_CAF1 54159 104 + 1 GCAAUAAAUACAUCAACGAUUCAUGAAUAUUUGAUAUAAGUAAUUGCAUUUUGUUAUGUGCCAAAUUAAAAUGAAAUGCCAUCAUCAUUUUUGAUGGCAUGUCA ((((((..((.(((((..(((....)))..))))).))..).)))))........................(((.(((((((((.......))))))))).))) ( -21.90) >DroPer_CAF1 43178 88 + 1 ---------------ACGAU-UAUGUAUAGUGGAUAUAAAUAAUGGCAUUUGGCUAUGUGCAAAAUUAAAAUGAAAUGCCAUCAUCAUUUUUGAUGCCAUGUCA ---------------.....-........((((.........(((((((((.((.....))............)))))))))(((((....))))))))).... ( -16.92) >consensus GCAAUAAAUACAUCAACGAUUCAUGAAUAUUUGAUAUAAAUAAUUGCAUUUUGUUAUGUGCCAAAUUAAAAUGAAAUGCCAUCAUCAUUUCUGAUGGCAUGUCA ........................................((((.((((........))))...))))...(((.(((((((((.......))))))))).))) (-15.75 = -16.08 + 0.33)


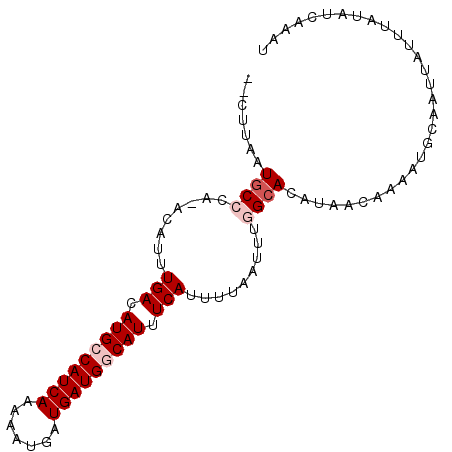
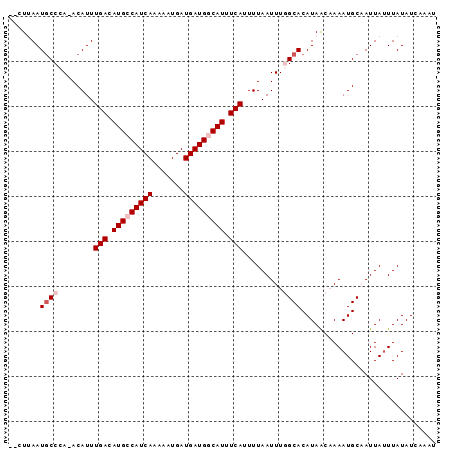
| Location | 17,090,015 – 17,090,107 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -15.35 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17090015 92 - 22407834 --CUUAAUGCCCA-ACAUUUGACAUGCCAUCAGAAAUGAUGAUGGCAUUUCAUUUUAAUUUGGCACAUAACAAAAUGCAAUUAUUUAUAUCAAAU --.....((((..-.....(((.(((((((((.......))))))))).))).........)))).............................. ( -20.69) >DroPse_CAF1 44799 95 - 1 GGCUCAAUGCCCAAACAUUUGACAUGGCAUCAAAAAUGAUGAUGGCAUUUCAUUUUAAUUUUGCACAUAGCCAAAUGCCAUUAUUUAUAUCCACU ((((...(((.........(((.(((.(((((.......))))).))).)))..........)))...))))....................... ( -19.11) >DroSec_CAF1 53759 92 - 1 --CUUAAUUCCCA-ACAUUUGACAUGCCAUCAGAAAUGAUGAUGGCAUUUCAUUUUAAUUUGGCACAUAACAAAAUGCAAUUAUUUAUAUCAAAU --........(((-(....(((.(((((((((.......))))))))).))).......))))................................ ( -17.80) >DroEre_CAF1 55146 92 - 1 --CUUAAUGCCCA-ACAUUUGACAUGCCAUCAGAAAUGAUGAUGGCAUUUCAUUUUAAUUUGGCACAUAACAAAAUGCAAUUAUUUAUAUCAAAU --.....((((..-.....(((.(((((((((.......))))))))).))).........)))).............................. ( -20.69) >DroYak_CAF1 54187 92 - 1 --CUUAAUGCCCA-ACAUUUGACAUGCCAUCAAAAAUGAUGAUGGCAUUUCAUUUUAAUUUGGCACAUAACAAAAUGCAAUUACUUAUAUCAAAU --.....((((..-.....(((.(((((((((.......))))))))).))).........)))).............................. ( -19.89) >DroPer_CAF1 43190 95 - 1 GGCUCAAUGCCCAAACAUUUGACAUGGCAUCAAAAAUGAUGAUGGCAUUUCAUUUUAAUUUUGCACAUAGCCAAAUGCCAUUAUUUAUAUCCACU ((((...(((.........(((.(((.(((((.......))))).))).)))..........)))...))))....................... ( -19.11) >consensus __CUUAAUGCCCA_ACAUUUGACAUGCCAUCAAAAAUGAUGAUGGCAUUUCAUUUUAAUUUGGCACAUAACAAAAUGCAAUUAUUUAUAUCAAAU .......((((........(((.(((((((((.......))))))))).))).........)))).............................. (-15.35 = -16.18 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:09 2006