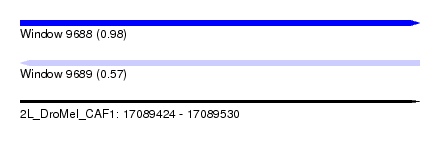
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,089,424 – 17,089,530 |
| Length | 106 |
| Max. P | 0.979090 |

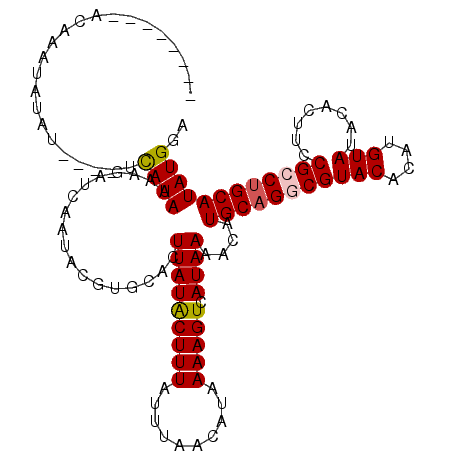
| Location | 17,089,424 – 17,089,530 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.16 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17089424 106 + 22407834 UCCAUAUGCAGGCGGAAGUGUAUACAUGUGUACGCCUGCAUGUUUUAUGACUUUUAUGUUAAAUAAAGUAUAAGUGCACGUAUUGAUAUUUUAUGA----AUAUAUUUGU-------- ...(((((((((((...(((....))).....)))))))))))......((...((((((..(((((((((.((((....)))).))))))))).)----)))))...))-------- ( -29.30) >DroSec_CAF1 53155 106 + 1 UCCAUAUGCAGGCGGAAGUGUAUACAUGUGUACGCCUGCAUGUUUUAUGACUUUUAUGCUAAAUAAAGUAUAAGUGCACGUAUUGAUCUUUUAUGA----AUAUAUUCGU-------- ...(((((((((((...(((....))).....)))))))))))....((...((((((((......))))))))..)).(((((.((.....)).)----))))......-------- ( -27.10) >DroSim_CAF1 48946 106 + 1 UCCAUAUGCAGGCGGAAGUGUAUACAUGUGUACGCCUGCAUGUUUUAUGACUUUUAUGCUAAAUAAAGUAUAAGUGCACGUAUUGAUCUUUUAUGA----AUAUAUUCGU-------- ...(((((((((((...(((....))).....)))))))))))....((...((((((((......))))))))..)).(((((.((.....)).)----))))......-------- ( -27.10) >DroEre_CAF1 54567 108 + 1 UCCAUAUGCAGGCGGAAGUGUGUACAUGUGUACGACUGCAUAUUUUAUGACUUUUAUGUUAAAUAAAGCAUAAGUGCACGUAUUGAUCUUUUAUGA----------GUAUAUACAUAU ...((((((((((....)).(((((....))))).))))))))..((((....(((((((......)))))))((((.((((.........)))).----------))))...)))). ( -26.50) >DroYak_CAF1 53628 117 + 1 UCCAUAUGC-GACGGAAGUGUAUACAUGUGUACGACUGCAUAUUUUAUUACUUUUAUGUUAAAUAAAGCAUAAGUGCACGUAUUGAUCUGAUAUACUUGUAUGUACUUGUAUACAUAU ....(((((-.((....))))))).((((((((((.(((..............(((((((......)))))))(((((.((((.........)))).)))))))).)))))))))).. ( -29.20) >consensus UCCAUAUGCAGGCGGAAGUGUAUACAUGUGUACGCCUGCAUGUUUUAUGACUUUUAUGUUAAAUAAAGUAUAAGUGCACGUAUUGAUCUUUUAUGA____AUAUAUUUGU________ ...(((((((((((...(((....))).....)))))))))))..((((...((((((((......))))))))....)))).................................... (-21.22 = -21.30 + 0.08)

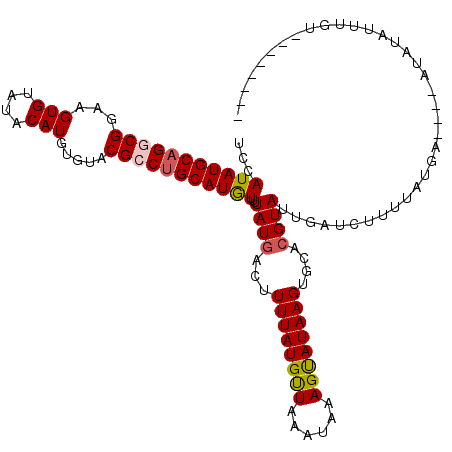
| Location | 17,089,424 – 17,089,530 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.16 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17089424 106 - 22407834 --------ACAAAUAUAU----UCAUAAAAUAUCAAUACGUGCACUUAUACUUUAUUUAACAUAAAAGUCAUAAAACAUGCAGGCGUACACAUGUAUACACUUCCGCCUGCAUAUGGA --------.........(----(((((............((....(((((((((..........))))).)))).)).(((((((((((....)))........)))))))))))))) ( -18.30) >DroSec_CAF1 53155 106 - 1 --------ACGAAUAUAU----UCAUAAAAGAUCAAUACGUGCACUUAUACUUUAUUUAGCAUAAAAGUCAUAAAACAUGCAGGCGUACACAUGUAUACACUUCCGCCUGCAUAUGGA --------.........(----(((((...(((......((((................))))....)))........(((((((((((....)))........)))))))))))))) ( -18.99) >DroSim_CAF1 48946 106 - 1 --------ACGAAUAUAU----UCAUAAAAGAUCAAUACGUGCACUUAUACUUUAUUUAGCAUAAAAGUCAUAAAACAUGCAGGCGUACACAUGUAUACACUUCCGCCUGCAUAUGGA --------.........(----(((((...(((......((((................))))....)))........(((((((((((....)))........)))))))))))))) ( -18.99) >DroEre_CAF1 54567 108 - 1 AUAUGUAUAUAC----------UCAUAAAAGAUCAAUACGUGCACUUAUGCUUUAUUUAACAUAAAAGUCAUAAAAUAUGCAGUCGUACACAUGUACACACUUCCGCCUGCAUAUGGA .(((((((....----------((......))...)))))))...(((((((((..........)))).))))).((((((((..((((....)))).(......).))))))))... ( -17.30) >DroYak_CAF1 53628 117 - 1 AUAUGUAUACAAGUACAUACAAGUAUAUCAGAUCAAUACGUGCACUUAUGCUUUAUUUAACAUAAAAGUAAUAAAAUAUGCAGUCGUACACAUGUAUACACUUCCGUC-GCAUAUGGA ...((((((((.((........((((.........))))(((((((((((.((((((..((......)))))))).)))).))).)))))).))))))))..(((((.-....))))) ( -21.60) >consensus ________ACAAAUAUAU____UCAUAAAAGAUCAAUACGUGCACUUAUACUUUAUUUAACAUAAAAGUCAUAAAACAUGCAGGCGUACACAUGUAUACACUUCCGCCUGCAUAUGGA .......................((((..................(((((((((..........))))).))))....(((((((((((....)))........)))))))))))).. (-12.74 = -12.94 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:07 2006