| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
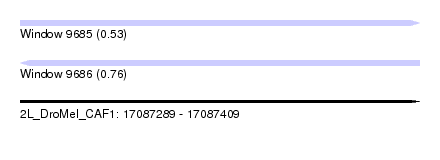
| Location | 17,087,289 – 17,087,409 |
| Length | 120 |
| Max. P | 0.763010 |

| Location | 17,087,289 – 17,087,409 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17087289 120 + 22407834 CGUCGGCCUUCUUUUUGCUGCAGUGGCAGUUGCAGUGGCAGUGGCAGUCGCAGUCGCAGUCGCAUUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAACAACCGGAAACUGACAGCAAA .(((((..((((..(((.(((((..((..(((..(((((.(((((.......))))).)))))....((((....))))...)))....))..))))).)))..)))).)))))...... ( -41.60) >DroPse_CAF1 42062 94 + 1 -UUUUGAUUUCUUUUCGCUGCAGUUGCAGUUGCAUUCGC-------------------------UUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAACAACCGGAAACUGACAGCAAA -...............((((((((((((((.((......-------------------------((..(((....)))..)).......)).))))))))....(....))).))))... ( -25.82) >DroSec_CAF1 51113 114 + 1 GGUCGGCCUUCUUUUUGCUGCAGUGGCAGUUGCAGUGGCAGUGGCAGUCG------CAGUCGCAUUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAAAAACCGGAAACUGACAGCAAA ((....)).....(((((((((((((...((((((..((.(((((.....------..)))))....((((....))))..........))..))))))...))....)))).))))))) ( -38.50) >DroSim_CAF1 46909 114 + 1 GGUCGGCCUUCUUUUUGCUGCAGUGGCAGUUGCAGUGGCAGUGGCAGUCG------CAGUCGCAUUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAACAACCGGAAACUGACAGCAAA ((....)).....(((((((((((((..(((((((..((.(((((.....------..)))))....((((....))))..........))..)))))))..))....)))).))))))) ( -41.20) >DroYak_CAF1 51542 114 + 1 GUUCGGCCUUUUUUUUGCUGCAGUGGCAGUGGCACUUGCAUUCGCAG------UCGCAGUCGCAUUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAACAACCGGAAACUGACAGCAAA .............(((((((((((.((((..((..((((....))))------..(((..(((....)))....)))............))..))))...........)))).))))))) ( -33.10) >DroPer_CAF1 40469 94 + 1 -UUUUGAUUUCUUUUCGCUGCAAUUGCAGUUGCAUUCGU-------------------------UUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAACAACCGGAAACUGACAGCAAA -...............((((((.(((((((.((....((-------------------------((.((((....))))))))......)).))))))).....(....))).))))... ( -24.10) >consensus GGUCGGCCUUCUUUUUGCUGCAGUGGCAGUUGCAGUGGCAGUGGCAG_________CAGUCGCAUUCGUGUUCAUGCAUAAACAAAAUGGCUGCUGCAACAACCGGAAACUGACAGCAAA .............((((((((((((((((((((....))............................((((....)))).........))))))))).......(....))).))))))) (-21.02 = -21.88 + 0.86)

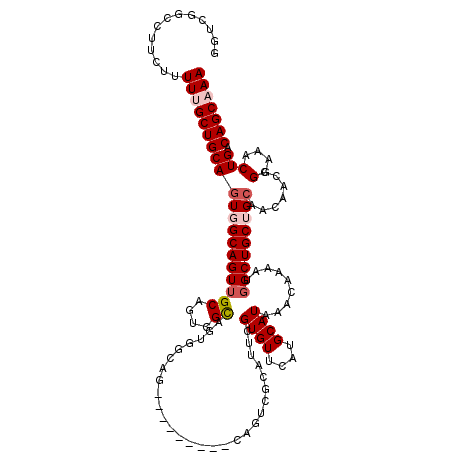

| Location | 17,087,289 – 17,087,409 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -23.39 |
| Energy contribution | -24.53 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17087289 120 - 22407834 UUUGCUGUCAGUUUCCGGUUGUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAAUGCGACUGCGACUGCGACUGCCACUGCCACUGCAACUGCCACUGCAGCAAAAAGAAGGCCGACG (((((((.((((....(((.((((((((((((((((((((((....)))))).))))).).))))...)))))).)))..(((....)))......)))))))))))............. ( -38.70) >DroPse_CAF1 42062 94 - 1 UUUGCUGUCAGUUUCCGGUUGUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAA-------------------------GCGAAUGCAACUGCAACUGCAGCGAAAAGAAAUCAAAA- (((((((.(((((....((.((((((...((..((.((((((....)))))).)).-------------------------))...)))))).))))))))))))))............- ( -28.20) >DroSec_CAF1 51113 114 - 1 UUUGCUGUCAGUUUCCGGUUUUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAAUGCGACUG------CGACUGCCACUGCCACUGCAACUGCCACUGCAGCAAAAAGAAGGCCGACC (((((((.((((....(((..((((((..((.((((((((((....)))))).))))))..)))------)))..)))..(((....)))......)))))))))))............. ( -35.10) >DroSim_CAF1 46909 114 - 1 UUUGCUGUCAGUUUCCGGUUGUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAAUGCGACUG------CGACUGCCACUGCCACUGCAACUGCCACUGCAGCAAAAAGAAGGCCGACC (((((((.((((....(((.(((((((..((.((((((((((....)))))).))))))..)))------)))).)))..(((....)))......)))))))))))............. ( -37.80) >DroYak_CAF1 51542 114 - 1 UUUGCUGUCAGUUUCCGGUUGUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAAUGCGACUGCGA------CUGCGAAUGCAAGUGCCACUGCCACUGCAGCAAAAAAAAGGCCGAAC (((((((.((((...((((.(((((((..((.((((((((((....)))))).))))))..))))))------)(((....)))......))))..)))))))))))............. ( -35.30) >DroPer_CAF1 40469 94 - 1 UUUGCUGUCAGUUUCCGGUUGUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAA-------------------------ACGAAUGCAACUGCAAUUGCAGCGAAAAGAAAUCAAAA- ((((((((..(((((.((((((....))))))....((((((....)))))).)))-------------------------))...(((....)))...))))))))............- ( -26.70) >consensus UUUGCUGUCAGUUUCCGGUUGUUGCAGCAGCCAUUUUGUUUAUGCAUGAACACGAAUGCGACUG_________CUGCCACUGCCACUGCAACUGCCACUGCAGCAAAAAGAAGGCCGAAC (((((((.((((....(((.((((((...(((((((((((((....)))))).))))).......................))...)))))).))))))))))))))............. (-23.39 = -24.53 + 1.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:04 2006