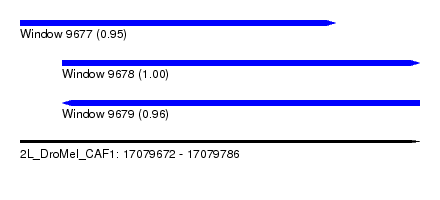
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,079,672 – 17,079,786 |
| Length | 114 |
| Max. P | 0.999723 |

| Location | 17,079,672 – 17,079,762 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17079672 90 + 22407834 GCUUCGC-----AUCCAUUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUA (..(((.-----..((....))..)))..)........(((((((((.((((((................))))))))).......))))))... ( -20.50) >DroSec_CAF1 43637 90 + 1 GCUUCGC-----AUCCAUUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUA (..(((.-----..((....))..)))..)........(((((((((.((((((................))))))))).......))))))... ( -20.50) >DroSim_CAF1 39138 90 + 1 GCUUCGC-----AUCCAUUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUA (..(((.-----..((....))..)))..)........(((((((((.((((((................))))))))).......))))))... ( -20.50) >DroEre_CAF1 44923 95 + 1 GCUUCGCCUCGCAUCCAUUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUA (..(((..(((........)))..)))..)........(((((((((.((((((................))))))))).......))))))... ( -20.10) >DroYak_CAF1 43949 90 + 1 GCUUCGC-----UUCCAUUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUA (..(((.-----..((....))..)))..)........(((((((((.((((((................))))))))).......))))))... ( -20.50) >consensus GCUUCGC_____AUCCAUUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUA (..(((........((....))..)))..)........(((((((((.((((((................))))))))).......))))))... (-17.30 = -17.30 + 0.00)

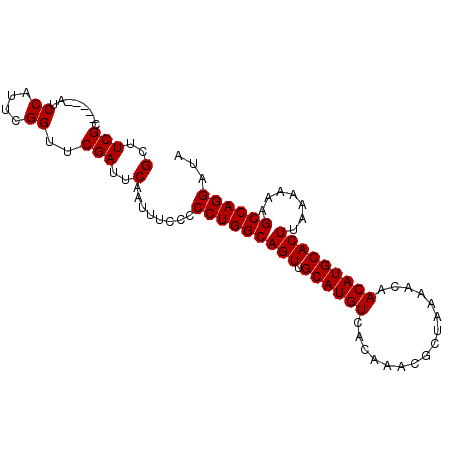
| Location | 17,079,684 – 17,079,786 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 98.63 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17079684 102 + 22407834 UUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUAUCUUCCUCAGUCCCCUGACAACCC ...((((.(............(((((((((.((((((................))))))))).......)))))).........((((....))))))))). ( -22.10) >DroSec_CAF1 43649 102 + 1 UUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUAUCUUCCUCAGUCCCCUGACAACCC ...((((.(............(((((((((.((((((................))))))))).......)))))).........((((....))))))))). ( -22.10) >DroSim_CAF1 39150 102 + 1 UUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUAUCUUCCUCAGUCCCCUGACAACCC ...((((.(............(((((((((.((((((................))))))))).......)))))).........((((....))))))))). ( -22.10) >DroEre_CAF1 44940 102 + 1 UUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUAUCUUCCUCGGCCCCCUGACAACCC ...((..(((...........(((((((((.((((((................))))))))).......)))))).........)))..))........... ( -21.55) >DroYak_CAF1 43961 102 + 1 UUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUAUCUUCCUCAGCCCCCUCACAACCC ...((((.((...........(((((((((.((((((................))))))))).......)))))).........))))))............ ( -19.05) >consensus UUCGGUUCGAUUCAAUUUCCCCCUGGCAGUUGCAUGUCACAAACGCUAAAACAACAUGCACUUAAAAAAGCCAGGAUAUCUUCCUCAGUCCCCUGACAACCC ...((((.(............(((((((((.((((((................))))))))).......)))))).........((((....))))))))). (-20.90 = -20.94 + 0.04)

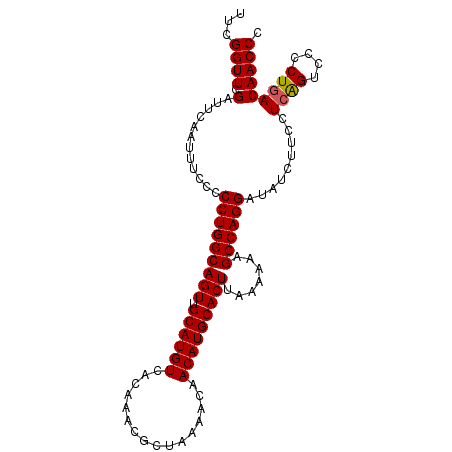
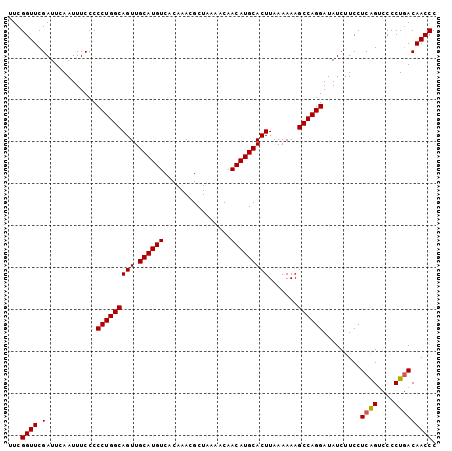
| Location | 17,079,684 – 17,079,786 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 98.63 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.17 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17079684 102 - 22407834 GGGUUGUCAGGGGACUGAGGAAGAUAUCCUGGCUUUUUUAAGUGCAUGUUGUUUUAGCGUUUGUGACAUGCAACUGCCAGGGGGAAAUUGAAUCGAACCGAA .((((.((((....))))........(((((((.......(((((((((..(..........)..)))))).)))))))))).............))))... ( -32.51) >DroSec_CAF1 43649 102 - 1 GGGUUGUCAGGGGACUGAGGAAGAUAUCCUGGCUUUUUUAAGUGCAUGUUGUUUUAGCGUUUGUGACAUGCAACUGCCAGGGGGAAAUUGAAUCGAACCGAA .((((.((((....))))........(((((((.......(((((((((..(..........)..)))))).)))))))))).............))))... ( -32.51) >DroSim_CAF1 39150 102 - 1 GGGUUGUCAGGGGACUGAGGAAGAUAUCCUGGCUUUUUUAAGUGCAUGUUGUUUUAGCGUUUGUGACAUGCAACUGCCAGGGGGAAAUUGAAUCGAACCGAA .((((.((((....))))........(((((((.......(((((((((..(..........)..)))))).)))))))))).............))))... ( -32.51) >DroEre_CAF1 44940 102 - 1 GGGUUGUCAGGGGGCCGAGGAAGAUAUCCUGGCUUUUUUAAGUGCAUGUUGUUUUAGCGUUUGUGACAUGCAACUGCCAGGGGGAAAUUGAAUCGAACCGAA .(((((((....)))(((....(((.(((((((.......(((((((((..(..........)..)))))).))))))))))....)))...)))))))... ( -28.71) >DroYak_CAF1 43961 102 - 1 GGGUUGUGAGGGGGCUGAGGAAGAUAUCCUGGCUUUUUUAAGUGCAUGUUGUUUUAGCGUUUGUGACAUGCAACUGCCAGGGGGAAAUUGAAUCGAACCGAA .((((.((((((((((.((((.....))))))))))))))(((((((((..(..........)..)))))).)))....................))))... ( -27.10) >consensus GGGUUGUCAGGGGACUGAGGAAGAUAUCCUGGCUUUUUUAAGUGCAUGUUGUUUUAGCGUUUGUGACAUGCAACUGCCAGGGGGAAAUUGAAUCGAACCGAA .((((.((((....))))........(((((((.......(((((((((..(..........)..)))))).)))))))))).............))))... (-28.77 = -29.17 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:57 2006