
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,078,110 – 17,078,210 |
| Length | 100 |
| Max. P | 0.742515 |

| Location | 17,078,110 – 17,078,210 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -13.61 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
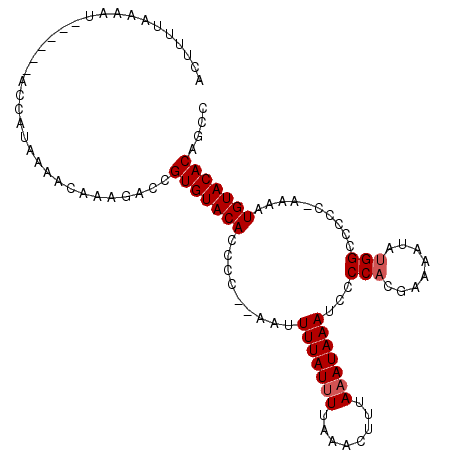
>2L_DroMel_CAF1 17078110 100 + 22407834 ACUUUUAAAAU------ACCAUAAAACAAAGGCCGUGUACACCC---AAUUUUAUUUUAAACUUUAAAUAAAUCCCCACGAAAAUAUGGCCCCC-AAAAUGUACACAGCC ...........------.............(((.(((((((...---...(((((((........)))))))...(((........))).....-....))))))).))) ( -16.40) >DroSim_CAF1 37537 103 + 1 ACUUUUAAAAU------ACCAUAAAACAAAGACCGUGUACACCCCCUAAGUUUAUUUUAAACUUUAAAUAAAUCCCCACGAAAAUAUGGCCCCC-AAAAUGUACACAGCC ...........------.................(((((((........((((((((........))))))))..(((........))).....-....))))))).... ( -12.90) >DroEre_CAF1 43317 107 + 1 GCUCUUAAAAUCAAAAGCCCGUAAAACAAAGCCCGUGUACACCCC---AUUUUAUAUUAAACUUUAAAUAAAUCCCCUCGAAAAUGUGGCCCCCAAAAAUGUACACAGCC (((.((......)).)))............((..(((((((((.(---(((((...........................)))))).))..........))))))).)). ( -11.53) >consensus ACUUUUAAAAU______ACCAUAAAACAAAGACCGUGUACACCCC__AAUUUUAUUUUAAACUUUAAAUAAAUCCCCACGAAAAUAUGGCCCCC_AAAAUGUACACAGCC ..................................(((((((.........(((((((........)))))))...(((........)))..........))))))).... ( -9.90 = -10.57 + 0.67)

| Location | 17,078,110 – 17,078,210 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17078110 100 - 22407834 GGCUGUGUACAUUUU-GGGGGCCAUAUUUUCGUGGGGAUUUAUUUAAAGUUUAAAAUAAAAUU---GGGUGUACACGGCCUUUGUUUUAUGGU------AUUUUAAAAGU ((((((((((((((.-.....((((......))))...(((((((........)))))))...---)))))))))))))).............------........... ( -25.80) >DroSim_CAF1 37537 103 - 1 GGCUGUGUACAUUUU-GGGGGCCAUAUUUUCGUGGGGAUUUAUUUAAAGUUUAAAAUAAACUUAGGGGGUGUACACGGUCUUUGUUUUAUGGU------AUUUUAAAAGU (((((((((((((((-..(((((((......))))...(((((((........))))))))))..))))))))))))))).............------........... ( -25.90) >DroEre_CAF1 43317 107 - 1 GGCUGUGUACAUUUUUGGGGGCCACAUUUUCGAGGGGAUUUAUUUAAAGUUUAAUAUAAAAU---GGGGUGUACACGGGCUUUGUUUUACGGGCUUUUGAUUUUAAGAGC (.((((((((((((.(((...)))((((((.....((((((.....)))))).....)))))---))))))))))))).)............(((((((....))))))) ( -25.10) >consensus GGCUGUGUACAUUUU_GGGGGCCAUAUUUUCGUGGGGAUUUAUUUAAAGUUUAAAAUAAAAUU__GGGGUGUACACGGCCUUUGUUUUAUGGU______AUUUUAAAAGU ((((((((((((((.......((((......))))...(((((((........)))))))......)))))))))))))).............................. (-20.60 = -21.27 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:54 2006