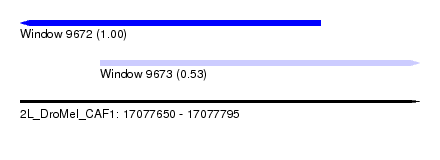
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,077,650 – 17,077,795 |
| Length | 145 |
| Max. P | 0.999131 |

| Location | 17,077,650 – 17,077,759 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 98.35 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
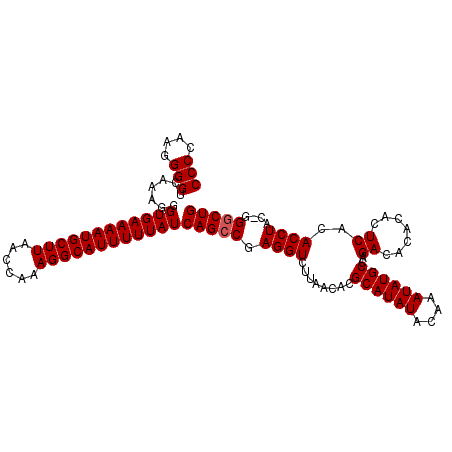
>2L_DroMel_CAF1 17077650 109 - 22407834 CCCCAAGGGGUCAAAGGGUGAAAAUGCUUAACCAAAGGCAUUUUUAUCAGCCGAGGUCUUAACACGCAUAUACAAAUAUGCCAGACACACACUCACACCUACCGGGCUG (((....)))......(((((((((((((......)))))))))))))((((.((((........((((((....))))))..((.......))..))))....)))). ( -31.40) >DroSec_CAF1 41577 108 - 1 CCCCAAGGGGUCAAAGAGUGAAAAUGCUUAACCAAAGGCAUUUUUAUCAGCCGAGGUCUUAACACGCAUAUACAAAUAUGCCAGACACACACUCACACCUAC-GGACUG (((....))).......((((((((((((......))))))))))))((((((((((........((((((....))))))..((.......))..)))).)-)).))) ( -29.90) >DroSim_CAF1 37064 108 - 1 CCCCAAGGGGUCAAAGGGUGAAAAUGCUUAACCAAAGGCAUUUUUAUCAGCCGAGGUCUUAACACGCAUAUACAAAUAUGCCAGACACACACUCACACCUAC-GGGCUG (((....)))......(((((((((((((......)))))))))))))((((.((((........((((((....))))))..((.......))..))))..-.)))). ( -32.00) >DroEre_CAF1 41326 109 - 1 CCCCAAGGGGUCAAAGGGUGAAAAUGCUUAACCAAAGGCAUUUUUAUCAGCCGAGGUCUUAACACGCAUAUACAAAUAUGCCAGACACACACUCACACCUACUGGGCUG .((((..((((.....(((((((((((((......)))))))))))))....(((..........((((((....))))))..........)))..))))..))))... ( -32.95) >DroYak_CAF1 41737 109 - 1 CCCCAAGGGGUCAAAGAGUGAAAAUGCUUAACCAAAGGCAUUUUUAUCAGCCGAGGUCUUAACACGCAUAUACAAAUAUGCCAGACACACACUCACACCUACUGGGCUG .((((..((((....((((((((((((((......)))))))))...........((((......((((((....)))))).))))...)))))..))))..))))... ( -31.50) >consensus CCCCAAGGGGUCAAAGGGUGAAAAUGCUUAACCAAAGGCAUUUUUAUCAGCCGAGGUCUUAACACGCAUAUACAAAUAUGCCAGACACACACUCACACCUAC_GGGCUG (((....))).......((((((((((((......))))))))))))(((((.((((........((((((....))))))..((.......))..))))....))))) (-28.32 = -28.52 + 0.20)

| Location | 17,077,679 – 17,077,795 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.43 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
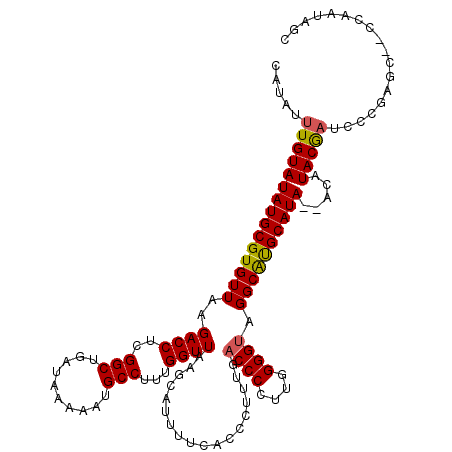
>2L_DroMel_CAF1 17077679 116 + 22407834 CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAGCAUUUUCACCCUUUGACCCCUUGGGGUAGGCAUGCAUA--ACAUACAAUCCCGAGC--CCAAUGGC .....(((((((((((((((..((((..(((..........)))...))))..........(((((..(....)..))))).))))))))))--...)))))......((--(....))) ( -31.30) >DroPse_CAF1 30303 115 + 1 CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAACAUUCUCACCCUUUGACCCCCUAGGG--GGCACGCAUACAUCGUACAUUCACAUUCAUUC---AAC ......((((((((((((....((((..(((..........)))...))))...................((((....))--))))))))).....))))).............---... ( -27.80) >DroSec_CAF1 41605 116 + 1 CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAGCAUUUUCACUCUUUGACCCCUUGGGGUAGGCGUGCAUA--ACAUACGAUCCCGAGU--CCAAUAGC .....(((((((((((((((...((((((((..........)))...(((((((...........)))))))....))))).))))))))))--...)))))........--........ ( -25.30) >DroSim_CAF1 37092 116 + 1 CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAGCAUUUUCACCCUUUGACCCCUUGGGGUAGGCAUGCAUA--ACAUACGAUCCCGAGC--CCAAUAGC .....(((((((((((((((..((((..(((..........)))...))))..........(((((..(....)..))))).))))))))))--...)))))........--........ ( -28.90) >DroEre_CAF1 41355 116 + 1 CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAGCAUUUUCACCCUUUGACCCCUUGGGGUAGGCAUGCAUA--ACAUACGAUCCUGAGC--CCAAUAGC ((...(((((((((((((((..((((..(((..........)))...))))..........(((((..(....)..))))).))))))))))--...)))))...))...--........ ( -29.10) >DroYak_CAF1 41766 118 + 1 CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAGCAUUUUCACUCUUUGACCCCUUGGGGUAGGCAUGCAUA--ACAUACGAUCCCGAGCCCCCAAUAGC .....(((((((((((((((...((((((((..........)))...(((((((...........)))))))....))))).))))))))))--...))))).................. ( -28.70) >consensus CAUAUUUGUAUAUGCGUGUUAAGACCUCGGCUGAUAAAAAUGCCUUUGGUUAAGCAUUUUCACCCUUUGACCCCUUGGGGUAGGCAUGCAUA__ACAUACGAUCCCGAGC__CCAAUAGC .....(((((((((((((((..((((..(((..........)))...))))..................((((....)))).)))))))))).....))))).................. (-26.60 = -26.43 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:52 2006