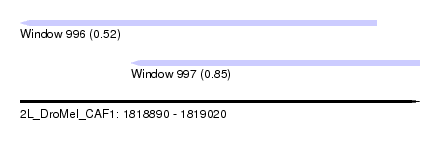
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,818,890 – 1,819,020 |
| Length | 130 |
| Max. P | 0.854498 |

| Location | 1,818,890 – 1,819,006 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
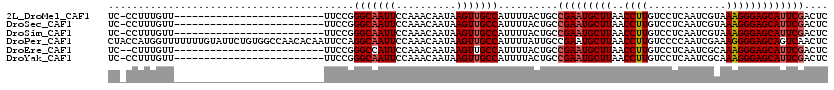
>2L_DroMel_CAF1 1818890 116 - 22407834 GGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUCCCGUUUUCCCUCGC----UUUUCCUCCAGAUGUUGCCCAA (((((((..(.(((.....))).............(((((((((..((((.............)))))))))))))..................----..........)..))))))).. ( -26.62) >DroSec_CAF1 38841 116 - 1 GGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUCCUGUCUUCCCUCGC----UUCUCCUCCAGAUGUAGCCCAA ((((.......(((.....))).........(((..((((((((..((((.............))))))))))))(((....))).........----........))).....)))).. ( -25.82) >DroSim_CAF1 16529 116 - 1 GGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUCCUGUUUUCCCUCGC----UUUUCCUCCAGAUGUUGCCCAA (((((((..........((((.........((.(.(((((((((..((((.............))))))))))))).)....))........))----))...........))))))).. ( -26.75) >DroPer_CAF1 30587 120 - 1 AGGCAAUUCCAAACAAUAAGUUGCCAUUUUAUUGCCGAAUGCUUAACCUUGUCCCCAAUCGAAAGGGGAGCAGUCAACUCCUGAGUUGUCCCGCCCCAUGCUCCCCCAGAUGUUGCCCAA .((((((..(...(((((((.......))))))).(((.((..............)).)))...(((((((((.(((((....))))).)........))))))))..)..))))))... ( -30.74) >DroEre_CAF1 27323 116 - 1 GGGCCAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGCAAAGGGAGCAUUCGACUCCUGUUCUCCCACGC----UUUUCCUCCAGAUGUUGCCCAA ((((((((.........((((.........((.(.(((((((((..((((((........)).))))))))))))).)....))........))----))........))))..)))).. ( -24.26) >DroYak_CAF1 3669 116 - 1 GGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGCAAAGGGAGCAUUCGACUCCUGUUUUCCCUCGC----UUUUCCUCCAGAUGUUGCCCAA (((((((..........((((.........((.(.(((((((((..((((((........)).))))))))))))).)....))........))----))...........))))))).. ( -27.43) >consensus GGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUCCUGUUUUCCCUCGC____UUUUCCUCCAGAUGUUGCCCAA (((((((..(.(((.....))).............(((((((((..((((.............)))))))))))))................................)..))))))).. (-22.91 = -23.60 + 0.70)


| Location | 1,818,926 – 1,819,020 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.95 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -19.89 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
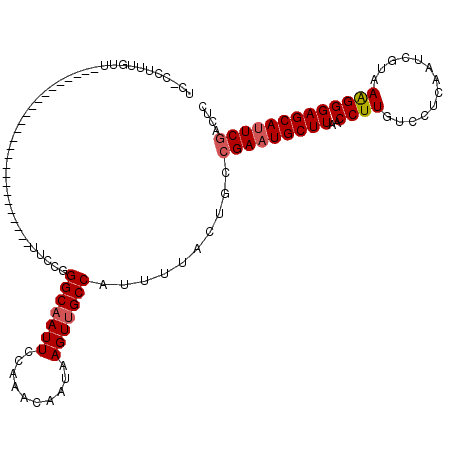
>2L_DroMel_CAF1 1818926 94 - 22407834 UC-CCUUUGUU-------------------------UUCCGGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUC ..-........-------------------------.....(((((((..........)))))))..........(((((((((..((((.............))))))))))))).... ( -21.82) >DroSec_CAF1 38877 94 - 1 UC-CCUUUGUU-------------------------UUCCGGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUC ..-........-------------------------.....(((((((..........)))))))..........(((((((((..((((.............))))))))))))).... ( -21.82) >DroSim_CAF1 16565 94 - 1 UC-CCUUUGUU-------------------------UUCCGGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUC ..-........-------------------------.....(((((((..........)))))))..........(((((((((..((((.............))))))))))))).... ( -21.82) >DroPer_CAF1 30627 120 - 1 CUACCAUGGUUUUUUUGUAUUCUGUGGCCAACACAAUUCCAGGCAAUUCCAAACAAUAAGUUGCCAUUUUAUUGCCGAAUGCUUAACCUUGUCCCCAAUCGAAAGGGGAGCAGUCAACUC .......((((.....((((((((((.....))))......((((((......(((....))).......))))))))))))..))))((((((((........))))).)))....... ( -29.92) >DroEre_CAF1 27359 93 - 1 UC--CUUUGUU-------------------------UUCCGGGCCAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGCAAAGGGAGCAUUCGACUC ..--..(((((-------------------------(...(((....)))))))))...................(((((((((..((((((........)).))))))))))))).... ( -18.40) >DroYak_CAF1 3705 94 - 1 UC-CCUUUGUU-------------------------UUCCGGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGCAAAGGGAGCAUUCGACUC ..-........-------------------------.....(((((((..........)))))))..........(((((((((..((((((........)).))))))))))))).... ( -22.50) >consensus UC_CCUUUGUU_________________________UUCCGGGCAAUUCCAAACAAUAAGUUGCCAUUUUACUGCCGAAUGCUUAACCUUGUCCUCAAUCGUAAAGGGAGCAUUCGACUC .........................................(((((((..........)))))))..........(((((((((..((((.............))))))))))))).... (-19.89 = -20.25 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:51 2006