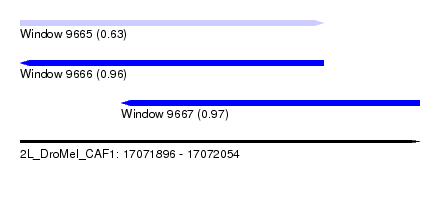
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,071,896 – 17,072,054 |
| Length | 158 |
| Max. P | 0.972078 |

| Location | 17,071,896 – 17,072,016 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -15.94 |
| Energy contribution | -17.90 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17071896 120 + 22407834 AAAAAGGCAAAAGCCAAGUGACACAGAAGCGACGGCAACAAAUUAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACUUCUGGGUGUUGCCACAAGCGGAAAAGCUGGUAGUGGGCAG ............(((..(..(((((((((.(.((...(((((((....(((......)))....)))).)))..))).)))))..))))..)(((.(((......)))....)))))).. ( -32.30) >DroPse_CAF1 23238 101 + 1 AAA-----------AAAGUGCCACAGAAGCGACGGCAACAAAUUAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACCUGGGGGAG--------GGCUGGGGUGUGGGGUGUGGGCAG ...-----------....((((...((.((....))............(((......))).))....((..(..(((((((.((....--------..)).)))))))..)..)))))). ( -28.90) >DroSec_CAF1 35856 120 + 1 AAAAAGGCAAAAGCCAAGUGACACAGAAGCGACGGCAACAAAUGAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACUUCUGGGUGUUGCCACAAGCGGAAAAGCUGGUAGUGGGCAG ............(((..(..(((((((((.(.((...((((((.....(((......))).....))).)))..))).)))))..))))..)(((.(((......)))....)))))).. ( -32.30) >DroEre_CAF1 35293 119 + 1 AAAAAGGCAAAAGCCAAGUGACACAGAAGCGACGGCAACAAAUUAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACUUCUGGGUGUUGCCACAAGGGG-AAAGCUGGUAGUGGGCGG ............(((..(..(((((((((.(.((...(((((((....(((......)))....)))).)))..))).)))))..))))..)(((..((..-....))....)))))).. ( -28.40) >DroYak_CAF1 35562 115 + 1 AAAAAGGCAAAAGCCAAGUGACACAGAAGCGACGGCAACAAAUUAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACUUCUGGGUGUUGCCACAAGG-----AGCUGCUACUGGAAGG .....(((....)))..(((.((((...((....))............(((......))).........)))).))).((((..(..((.((.......-----.)).))..)..)))). ( -29.00) >DroPer_CAF1 21190 93 + 1 AAA-----------AAAGUGCCACAGAAGCGACGGCAACAAAUUAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACCUGGGGA-G--------GG-------GUGGGGUGUGGGCAG ...-----------....(((((((...((....))............(((......))).........)))..(((((((.(...-.--------..-------.).))))))))))). ( -23.40) >consensus AAAAAGGCAAAAGCCAAGUGACACAGAAGCGACGGCAACAAAUUAAAAGCCAUAAAAGGCAUCAAAUUAUGUGACGCACUUCUGGGUGUUGCCACAAGCGG_AAAGCUGGUAGUGGGCAG .....(((((.(.(((((((.((((...((....))............(((......))).........))))...))))..))).).)))))........................... (-15.94 = -17.90 + 1.96)

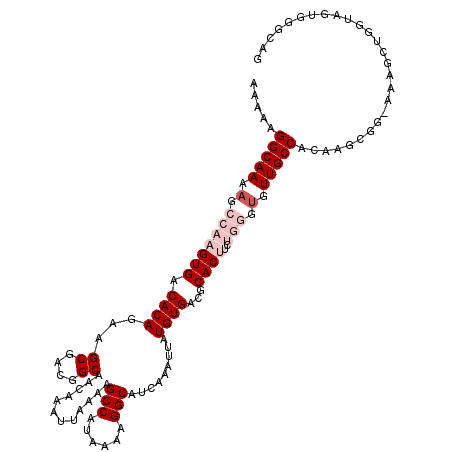
| Location | 17,071,896 – 17,072,016 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -19.18 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17071896 120 - 22407834 CUGCCCACUACCAGCUUUUCCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGCUUCUGUGUCACUUGGCUUUUGCCUUUUU .((((.((....(((......))).))))))..((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))))))).)).....(((....)))..... ( -34.70) >DroPse_CAF1 23238 101 - 1 CUGCCCACACCCCACACCCCAGCC--------CUCCCCCAGGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGCUUCUGUGGCACUUU-----------UUU ...........(((((....(((.--------.......(((.((((((.......)))))))))..(((((............))))).)))..)))))......-----------... ( -19.70) >DroSec_CAF1 35856 120 - 1 CUGCCCACUACCAGCUUUUCCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUCAUUUGUUGCCGUCGCUUCUGUGUCACUUGGCUUUUGCCUUUUU .((((.((....(((......))).))))))..((.((((((((((.((.((((..(((.(((......)))...))).)))))).)).)))))))).)).....(((....)))..... ( -34.50) >DroEre_CAF1 35293 119 - 1 CCGCCCACUACCAGCUUU-CCCCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGCUUCUGUGUCACUUGGCUUUUGCCUUUUU .............((...-..((..((((((.....((((((((((.((.((((.((((.(((......)))..)))).)))))).)).))))))))))))))..)).....))...... ( -33.10) >DroYak_CAF1 35562 115 - 1 CCUUCCAGUAGCAGCU-----CCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGCUUCUGUGUCACUUGGCUUUUGCCUUUUU .......((((.((((-----....((((((.....((((((((((.((.((((.((((.(((......)))..)))).)))))).)).))))))))))))))..)))).))))...... ( -34.80) >DroPer_CAF1 21190 93 - 1 CUGCCCACACCCCAC-------CC--------C-UCCCCAGGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGCUUCUGUGGCACUUU-----------UUU .((((.(((....((-------(.--------.-......)))(((.(.(((((.((((.(((......)))..)))).))).))..).)))...)))))))....-----------... ( -19.30) >consensus CUGCCCACUACCAGCUUU_CCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGCUUCUGUGUCACUUGGCUUUUGCCUUUUU .................................((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))))))).)).....(((....)))..... (-19.18 = -20.10 + 0.92)

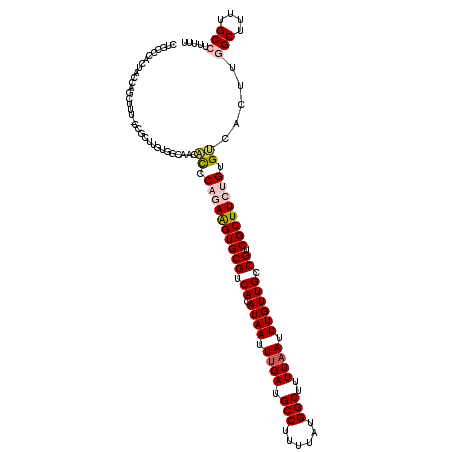
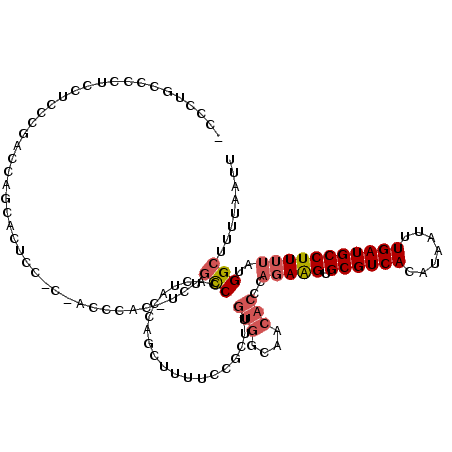
| Location | 17,071,936 – 17,072,054 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -14.29 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17071936 118 - 22407834 -CCCUUCUCCACCUUCCGACCUCCACUCCACCACCCAU-UCUGCCCACUACCAGCUUUUCCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUU -.....................................-...(((..((((.(((......))).))))........(((((.((((((.......)))))))))))..)))........ ( -20.60) >DroPse_CAF1 23267 106 - 1 CUCCUUCUG---UUUGUGGC-UGGAAACA-CCCCUCCC-CCUGCCCACACCCCACACCCCAGCC--------CUCCCCCAGGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUU .........---..((((((-((....))-........-......................(((--------((.....))).))))))))....((((.(((......)))..)))).. ( -19.10) >DroSec_CAF1 35896 115 - 1 -CCCUGCCCCUCCUCACGACCAGCACUCC---ACCCAU-UCUGCCCACUACCAGCUUUUCCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUCAUU -................(((..(((((..---......-((((.........(((......))).(((....))).))))))))))))........(((.(((......)))...))).. ( -22.50) >DroSim_CAF1 31191 118 - 1 -CCCUGCCCCUCCUCACGACCAGCACUCCACCACCCAU-UCUGCCCACUACCAGCUUUUCCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUU -....(((..............................-..((((.((....(((......))).))))))......(((((.((((((.......)))))))))))..)))........ ( -21.90) >DroEre_CAF1 35333 107 - 1 --CGCGCCCC--CACCCCAUCAGC-CCCU------CAC-UCCGCCCACUACCAGCUUU-CCCCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUU --........--.........(((-(...------...-...................-......(((....)))..(((((.((((((.......)))))))))))..))))....... ( -19.20) >DroYak_CAF1 35602 109 - 1 -UCGUGCCCCUUUACCCCAUCAAC-CACCAC----CACCCCCUUCCAGUAGCAGCU-----CCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUU -....(((................-......----.....................-----....(((....)))..(((((.((((((.......)))))))))))..)))........ ( -18.40) >consensus _CCCUGCCCCUCCUCCCGACCAGCACUCC_C_ACCCAC_UCUGCCCACUACCAGCUUUUCCGCUUGUGGCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUU ..........................................(((....................(((....)))..(((((.((((((.......)))))))))))..)))........ (-14.29 = -15.02 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:45 2006