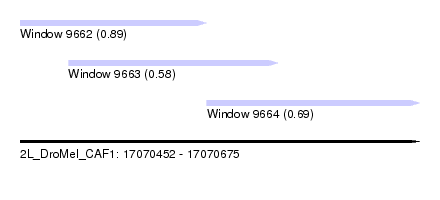
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,070,452 – 17,070,675 |
| Length | 223 |
| Max. P | 0.891376 |

| Location | 17,070,452 – 17,070,556 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17070452 104 + 22407834 CCAU------AUACAAACUCUGCGAAAUACCCAUCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC-CGCA-CAUGGCACAAAAGGUUGG ((..------..........(((((........))))).....((((.((((((((.........))))))))..)))).......((-((..-..))))......)).... ( -19.20) >DroSec_CAF1 34430 104 + 1 CCAU------AUACCAACUCGGCGAAAUACCCAUCGCACACAUUUUACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC-CGCA-CAUGGCACAAAAGGUUGG ....------...((((((..((((........))))...........((((((((.........)))))))).............((-((..-..))))......)))))) ( -23.20) >DroSim_CAF1 29725 106 + 1 CCAU------AUACCAACUCGGCGAAAUACCCAUCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGCCCGCAUCAUGGCACAAAAGGUUGG ....------...((((((..((((........))))......((((.((((((((.........))))))))..)))).......(((........)))......)))))) ( -23.20) >DroEre_CAF1 33929 110 + 1 ACAUACUCGUAUACCAACUCUGCGAUGGGUAUUUCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGAAGC-CGCA-CAUGGCACAAAAGGUUGG .............((((((.(((((........))))).....((((.((((((((.........))))))))..)))).......((-((..-..))))......)))))) ( -25.20) >DroYak_CAF1 33332 104 + 1 ACAU------AUACCAACUCUGCGGAAUACCCAUCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC-CGCA-CAUGGCACAAAAGGUUGC ....------....(((((.(((((.....))...........((((.((((((((.........))))))))..))))....)))((-((..-..))))......))))). ( -21.00) >consensus CCAU______AUACCAACUCUGCGAAAUACCCAUCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC_CGCA_CAUGGCACAAAAGGUUGG .............((((((..((((........))))......((((.((((((((.........))))))))..))))...........((......))......)))))) (-18.22 = -18.30 + 0.08)

| Location | 17,070,479 – 17,070,596 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17070479 117 + 22407834 -UCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC-CGCA-CAUGGCACAAAAGGUUGGCCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGG -...(((.((((((((((((((((.........))))))))..........((...-.)).-..((((.(((....)))))))...............))))))))..........))). ( -25.60) >DroPse_CAF1 22167 90 + 1 CUUGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUA--------------------CAAAAGGUUGGCCAACAAAAACGCAAAUGG--AAAAGCAAAA-A------- .((((...((((((..((((((((.........))))))))........--------------------.....((....)).........).))))).--....))))..-.------- ( -15.50) >DroSec_CAF1 34457 117 + 1 -UCGCACACAUUUUACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC-CGCA-CAUGGCACAAAAGGUUGGCCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGG -.....(((.......((((((((.........)))))))).(((......(((..-..((-(.((((.(((....))))))).......(......))))...)))......)))))). ( -25.30) >DroSim_CAF1 29752 119 + 1 -UCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGCCCGCAUCAUGGCACAAAAGGUUGGCCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGG -...(((.((((((((((((((((.........))))))))..........((.....))....((((.(((....)))))))...............))))))))..........))). ( -25.60) >DroEre_CAF1 33962 116 + 1 -UCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGAAGC-CGCA-CAUGGCACAAAAGGUUGGCCAACAAAAACUCCACUGGUGAAAUGCAAA-AGCAGGUGG -.....((((((((((((((((((.........))))))))...............-....-..((((.(((....)))))))...............)))))))((...-.))..))). ( -26.10) >DroPer_CAF1 20130 90 + 1 CUUGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUA--------------------CAAAAGGUUGGCCAACAAAAACGCAAAUGG--AAAAGCAAAA-A------- .((((...((((((..((((((((.........))))))))........--------------------.....((....)).........).))))).--....))))..-.------- ( -15.50) >consensus _UCGCACACAUUUCACGCAUUUUGAUAUGCAAACAAAAUGCACUGAAUAAAGCAGC_CGCA_CAUGGCACAAAAGGUUGGCCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGG ........((((((((((((((((.........)))))))).................................((....))................)))))))).............. (-15.02 = -15.93 + 0.92)


| Location | 17,070,556 – 17,070,675 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17070556 119 + 22407834 CCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGGGGUAGGUAACAAAAUAAAAAACGUAUAGAACGAAUGGUAAAAACCGUCGAAAUUUGCACGUGAACACAAAGAAGCAAUU .........((((((((.((............)).)))))))).................((((.((((.(((.((((....)))))))...)))).)))).................. ( -21.10) >DroSec_CAF1 34534 119 + 1 CCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGGGGUAGGUAACAAAAUAAAAAACGUAUAGAACGAAUGGUAAAAACCGUCGAAAUUUGCACGUGAACACAAAGAAGCAAUU .........((((((((.((............)).)))))))).................((((.((((.(((.((((....)))))))...)))).)))).................. ( -21.10) >DroSim_CAF1 29831 119 + 1 CCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGGGGUAGGUAACAAAAUAAAAAACGUAUAGAACGAAUGGUAAAAACCGUCGAAAUUUGCACGUGAACACAAAGAAGCAAUU .........((((((((.((............)).)))))))).................((((.((((.(((.((((....)))))))...)))).)))).................. ( -21.10) >DroYak_CAF1 33436 118 + 1 CCAACAAAAACUCCACUGGUGAAAUGCAAA-AACAGGUGGGGUAGGUAACAAAAUAAAAAACGUAUAGAACGAAUGGUAAAAACCGUCGAAAUUUGCACGUGAACACAAAGAAGCAAUU .........((((((((.((..........-.)).)))))))).................((((.((((.(((.((((....)))))))...)))).)))).................. ( -21.20) >consensus CCAACAAAAACUCCACUGGUGAAAUGCAAAAAACAGGUGGGGUAGGUAACAAAAUAAAAAACGUAUAGAACGAAUGGUAAAAACCGUCGAAAUUUGCACGUGAACACAAAGAAGCAAUU .........((((((((.((............)).)))))))).................((((.((((.(((.((((....)))))))...)))).)))).................. (-21.10 = -21.10 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:42 2006